













| General Information: |
|
| Name(s) found: |
DNM1
[HGNC (HUGO)]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 864 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGNRGMEDLI PLVNRLQDAF SAIGQNADLD LPQIAVVGGQ SAGKSSVLEN FVGRDFLPRG 60
61 SGIVTRRPLV LQLVNATTEY AEFLHCKGKK FTDFEEVRLE IEAETDRVTG TNKGISPVPI 120
121 NLRVYSPHVL NLTLVDLPGM TKVPVGDQPP DIEFQIRDML MQFVTKENCL ILAVSPANSD 180
181 LANSDALKVA KEVDPQGQRT IGVITKLDLM DEGTDARDVL ENKLLPLRRG YIGVVNRSQK 240
241 DIDGKKDITA ALAAERKFFL SHPSYRHLAD RMGTPYLQKV LNQQLTNHIR DTLPGLRNKL 300
301 QSQLLSIEKE VEEYKNFRPD DPARKTKALL QMVQQFAVDF EKRIEGSGDQ IDTYELSGGA 360
361 RINRIFHERF PFELVKMEFD EKELRREISY AIKNIHGIRT GLFTPDMAFE TIVKKQVKKI 420
421 REPCLKCVDM VISELISTVR QCTKKLQQYP RLREEMERIV TTHIREREGR TKEQVMLLID 480
481 IELAYMNTNH EDFIGFANAQ QRSNQMNKKK TSGNQDEILV IRKGWLTINN IGIMKGGSKE 540
541 YWFVLTAENL SWYKDDEEKE KKYMLSVDNL KLRDVEKGFM SSKHIFALFN TEQRNVYKDY 600
601 RQLELACETQ EEVDSWKASF LRAGVYPERV GDKEKASETE ENGSDSFMHS MDPQLERQVE 660
661 TIRNLVDSYM AIVNKTVRDL MPKTIMHLMI NNTKEFIFSE LLANLYSCGD QNTLMEESAE 720
721 QAQRRDEMLR MYHALKEALS IIGDINTTTV STPMPPPVDD SWLQVQSVPA GRRSPTSSPT 780
781 PQRRAPAVPP ARPGSRGPAP GPPPAGSALG GAPPVPSRPG ASPDPFGPPP QVPSRPNRAP 840
841 PGVPSRSGQA SPSRPESPRP PFDL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
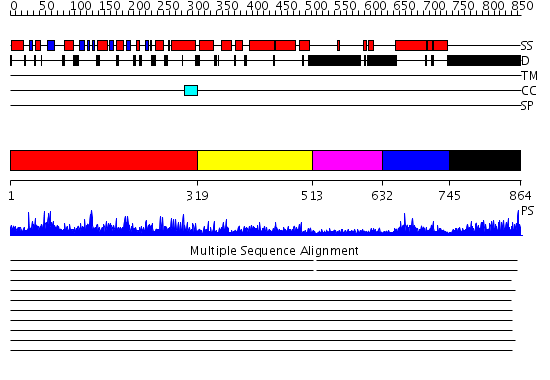
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..318] | 136.0 | Structure of the nucleotide-free myosin II motor domain from Dictyostelium discoideum fused to the GTPase domain of dynamin 1 from Rattus norvegicus | |
| 2 | View Details | [319..512] | 1.22296 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [513..631] | 30.39794 | Dynamin | |
| 4 | View Details | [632..744] | 28.769551 | No description for PF02212.9 was found. Confident ab initio structure predictions are available. | |
| 5 | View Details | [745..864] | 3.522879 | No description for 1y0fA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)