













| General Information: |
|
| Name(s) found: |
chn-PC /
FBpp0110305
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1214 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC]
intracellular [IEA] |
| Biological Process: |
negative regulation of transcription from RNA polymerase II promoter
[NAS]
eye development [IMP] embryonic development via the syncytial blastoderm [IMP] nervous system development [NAS] peripheral nervous system development [IMP] negative regulation of compound eye cone cell fate specification [IMP] positive regulation of transcription from RNA polymerase II promoter [IMP] progression of morphogenetic furrow during compound eye morphogenesis [IMP] sensory organ development [IMP] |
| Molecular Function: |
transcription factor activity
[ISS][NAS]
zinc ion binding [IEA] protein binding [IPI] RNA polymerase II transcription factor activity, enhancer binding [IDA] sequence-specific DNA binding [IDA] specific transcriptional repressor activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATLIPVNGG HPAASGQSSN VEATYEDMFK EITRKLYGEE TGNGLHTLGT PVAQVATSGP 60
61 TAVPEGEQRS FTNLQQLDRS AAPSIEYESS AAGASGNNVA TTQANVIQQQ QQQQQQAESG 120
121 NSVVVTASSG ATVVPAPSVA AVGGFKSEDH LSTAFGLAAL MQNGFAAGQA GLLKAGEQQQ 180
181 RWAQDGSGLV AAAAAEPQLV QWTSGGKLQS YAHVNQQQQQ QQQPHQSTPK SKKHRQEHAA 240
241 ELIYASPSTS ANAAQNLAQS TPTSAPSNSS GGSTSSSGGG GGRKKAAQAA AAAAAANGVH 300
301 IQKRYACTHC PYSTDRRDLY TRHENIHKDE KPFQCYACLK QFNRADHVKK HFLRMHRELQ 360
361 YDINKTRRHV SAGSGSSGSG SSGSGSHHSG GRGNVTINSA GVNIDNAFLE AQRHPTSSSM 420
421 SIVETIEAVA SATDMPLAQL KQEKMDDGAG VVLPLHVGVM QQPVASSSSG SSGSHGGNGN 480
481 GGSGSGLLKP KREKRFTCCY CPWSGADKWG LKRHLNTHTK PFVCLLCDYK AARSERLATH 540
541 VLKVHNKRAC SKCSYLADTQ EEYQAHMSDV HGNVGNANGG GGAVTIYTTT TNEGVAGGGG 600
601 GGGGGISGNI SGGGPLQEII VNPSSMVGWR LSANGSLIPP HDLLTGGLPN AATQKRGSER 660
661 LFQYLEAEGS DPEDYARLLK MDAISRNTAS VAQDFHKAGG VHELKIPANH QLLFNNKLPS 720
721 QWTTREAAAL LYSLSNMGGG SASSVSGSQR QKFGMRARQH STGEDDENTP SSASSSSFSG 780
781 DEFNMSATSP LKLSRHAIKL EKMDEMDAKD MGPTKAMMAT AFLEAANYEQ TAIELLASKR 840
841 KIKVENDNDE DQENQQHQPH QQHHSQQQQQ QRLQLIKSSP AYKLNNNNNN NSNNNNYYKD 900
901 KTSHRNAVHH HRQDDKENNK TKSPGTAAVS VAAAAATSPP SISGPSNQTP FLTQMEYQNL 960
961 NRIGTQFQNY VKDIINKYYA AETPLMLAAA AAALPTATTT GQQQQPELDI ENLSPSKRRR1020
1021 LLSETEEYIE YLRNKEDITL TIAPKVQPPA PVTSLLKRQL DLSTPRRSPK KAAPAHSNSA1080
1081 SNASRKSLNQ LATLLPLLAD AASQQEYLAA PLDFSKKSSS RKQAQPKKIR LTPEAVVTLL1140
1141 RDKYLNRMVR QRLGCLKCNQ LRKNSSISFN YHTLGSLALH KYWRHGRLGS SSTRREKLQA1200
1201 ALQKRISRGQ ADKC |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
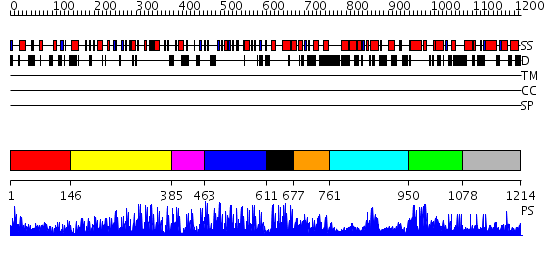
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..145] | 2.434989 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [146..384] | 48.522879 | No description for 2i13A was found. | |
| 3 | View Details | [385..462] | 16.0 | Five-finger GLI1 | |
| 4 | View Details | [463..610] | 17.0 | Transcription factor IIIA, TFIIIA | |
| 5 | View Details | [611..676] | 3.69897 | HIGH-RESOLUTION SOLUTION STRUCTURE OF THE DOUBLE CYS2*HIS2 ZINC FINGER FROM THE HUMAN ENHANCER BINDING PROTEIN MBP-1 | |
| 6 | View Details | [677..760] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [761..949] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [950..1077] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1078..1214] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)