













| General Information: |
|
| Name(s) found: |
PIF1_SCHPO
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 805 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFSCQSLYKF SHSFRKRIPV MFQRAQQKSS LLHTQNESSH QPSLNKLGGF SSASLNFNSS 60
61 RSSTNDDQQT FSSQSDNLPS SPITLPAKRG RSAASLKQLD NTVGFDVSKP SLPVFENSGL 120
121 GSKYSTEIAN GVYIDENDFD DDLLLENDID QKPIPWSSSP IEHTKLTKSM LSSEKRSKNH 180
181 LSKIYEDHTS EKGASSVISS NITRQGIKRS RTLPWAVDPY RYGDPDPKRT STSADISQHT 240
241 VSNDSSNKLS NGRSSSLDSL AKKRMSKSKS TPQISKKFSV PLNSASKSPI GSSLFKTSDS 300
301 RKKSVPSIFL SDEQKRILDM VVEQQHSIFF TGSAGTGKSV LLRKIIEVLK SKYRKQSDRV 360
361 AVTASTGLAA CNIGGVTLHS FAGVGLARES VDLLVSKIKK NKKCVNRWLR TRVLIIDEVS 420
421 MVDAELMDKL EEVARVIRKD SKPFGGIQLV LTGDFFQLPP VPENGKESKF CFESQTWKSA 480
481 LDFTIGLTHV FRQKDEEFVK MLNELRLGKL SDESVRKFKV LNRTIEYEDG LLPTELFPTR 540
541 YEVERSNDMR MQQINQNPVT FTAIDSGTVR DKEFRDRLLQ GCMAPATLVL KVNAQVMLIK 600
601 NIDDQLVNGS LGKVIGFIDD ETYQMEKKDA EMQGRNAFEY DSLDISPFDL PDVKQKKYKL 660
661 IAMRKASSTA IKWPLVRFKL PNGGERTIVV QRETWNIELP NGEVQASRSQ IPLILAYAIS 720
721 IHKAQGQTLD RVKVDLGRVF EKGQAYVALS RATTQEGLQV LNFSPAKVMA HPKVVQFYKQ 780
781 LASVNGLPIR NENKAPVQMR GVKNK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
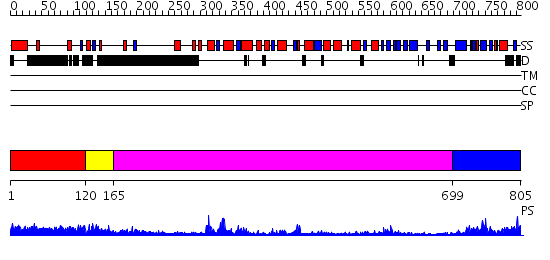
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..119] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [120..164] | 44.221849 | No description for 2gjkA was found. | |
| 3 | View Details | [165..698] | 28.221849 | RecBCD:DNA complex | |
| 4 | View Details | [699..805] | 62.69897 | No description for 2is1A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)