













| General Information: |
|
| Name(s) found: |
gi|78707559
[NCBI NR]
gi|71911696 [NCBI NR] |
| Description(s) found:
Found 5 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1196 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Z disc
[IDA]
muscle tendon junction [IDA] |
| Biological Process: | NONE FOUND |
| Molecular Function: |
zinc ion binding
[IEA]
protein binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAQPQLLQIK LSRFDAQPWG FRLQGGTDFA QPLLVQKVNA GSLSEQAGLQ PGDAVVKIND 60
61 VDVFNLRHKD AQDIVVRSGN NFVITVQRGG STWRPHVTPT GNVPQPNSPY LQTVTKTSLA 120
121 HKQQDSQHIG CGYNNAARPF SNGGDGGVKS IVNKQYNTPV GIYSDESIAE TLSAQAEVLA 180
181 GGVLGVNFKK NEKEYQGDRS EVLKFLREEE TGQSTPAFGN SHYEHDAPQQ LQQPQQQYNQ 240
241 HQQHYHQQQQ QQQSSTTRHV SAPVNSPKPP STGGLPTGQN ICTECERLIT GVFVRIKDKN 300
301 LHVECFKCAT CGTSLKNQGY YNFNNKLYCD IHAKQAAINN PPTGTEGYVP VPIKPNTKLS 360
361 ASTISSALNS HGYGGHSNGY SNGNSTPAPA PPESESSQEL PLPPPPSPTQ LLQYEAEQQV 420
421 LPEPHMSTIQ QPQQQQKLEH TRTHSSLSSI SSGSSSSGAQ SVTNYRLQAA QNENDMNTQN 480
481 KSPNAYNQLL KEYSNKLQQQ HHHNTTQHTT ATTAPSLKHN NTAKPFAVAA TSTPQQHLPH 540
541 QQHQQQPLVA ALTATLANQL KFNPHQVASS QATATVATVA PSAATAATAA ATPQAATATD 600
601 SPAATASSSD NMSAYVADEP SSIYGQISAE SVALAPPPPQ PPTAGGGDQP FEYVTLTGNV 660
661 IRSVQAPGKG ACPSYKVNQG YARPFGAAAP KSPVSYPPQQ QQQSPRPAPG GQNPYATLPR 720
721 SNVGQQVPTG AQSAAKSTNL HASPQTFATL PRPHPQATDI PAQELQKDNQ LESDATSMLA 780
781 ESVGKINMGD KTQAFMQEAG KIITNMLEAR LANNGGQNGP LSHAGSSLSL RSNSSSNLSK 840
841 SPMIVRKRLD LDDFKNLDPT QPVALQPVIA VHSQKDLPTE VGNQKAQPQA PRLEQTLQME 900
901 LAAQHPMGKI LDLCDGGKAI DTPEPVAFRN NLKRTGATHA ADRRSYIEPK QGAINATSVA 960
961 TQNGNQNGGQ AGNQNATQGS APTYSVSVKA LGPHSEDQTT MSEENERAVS QLLKEGKRPV1020
1021 CCQCNKEITS GPFITALGRI WCPDHFICVN GNCRRPLQDI GFVEEKGDLY CEYCFEKYLA1080
1081 PTCSKCAGKI KGDCLNAIGK HFHPECFTCG QCGKIFGNRP FFLEDGNAYC EADWNELFTT1140
1141 KCFACGFPVE AGDRWVEALN HNYHSQCFNC TFCKQNLEGQ SFYNKGGRPF CKNHAR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
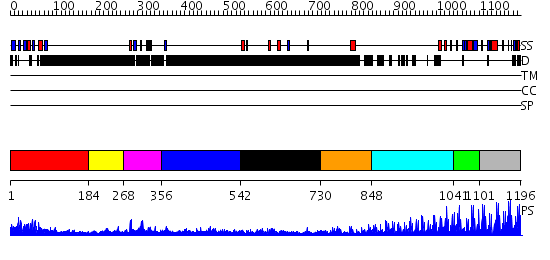
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..183] | 24.522879 | Solution structure of PDZ domain of mouse Alpha-actinin-2 associated LIM protein | |
| 2 | View Details | [184..267] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [268..355] | 7.0 | Solution structure of the LIM domain of alpha-actinin-2 associated LIM protein | |
| 4 | View Details | [356..541] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [542..729] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [730..847] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [848..1040] | 35.045757 | No description for 2i13A was found. | |
| 8 | View Details | [1041..1100] | 1.74 | Cysteine-rich (intestinal) protein, CRP, CRIP | |
| 9 | View Details | [1101..1196] | 15.39794 | Ying-yang 1 (yy1, zinc finger domain) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)