













| General Information: |
|
| Name(s) found: |
alp-1 /
CE39401
[WormBase]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 684 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
pharyngeal pumping
[IMP locomotory behavior [IMP determination of adult lifespan [IMP |
| Molecular Function: |
zinc ion binding
[IEA protein binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MERVTVRMAR SDRRTPWGFG VTEAPDGAVL IVNVIGGSLA DRAGLRNGDV VDRLEDLDNL 60
61 DINAVDRLLV TAHEKIELIV TRQPSGAATR IWRPEVTENT GPGQDQTPYR VNLQHSSDTR 120
121 PPQGFNNSAL PFETDQRVKH MQYNSPLGIY SDKSAAEQYV QQTQGLGDNS GARAAAQRQD 180
181 EPAYLRSETL RLLKEQEHGS AGGAGTTLSN HPNPSGLPVC FMCTRPILGV MARAAGKNLH 240
241 GDCLSCATCG NSLRNVGHHF IEDKFYCDIH GTQRKHGGRP GMDPNIFVKS SPTAAQPVHI 300
301 QDSPRAAYHP QVNTARPVSV SPAPSAGSKT VTTHYHTPKT QGILSPGQRG VPTAAFGADL 360
361 SKSVTYGGGV GPGGTHYDSN RLSPAPSQYS TQSRNQSQQH YQQHVVQAPP PVPSNPPPSN 420
421 DTTAEDSRNF LASRLGQVMI DENKKAGGSN GSSQHHNQQG SSYTKTSFER TTTETRPQQN 480
481 TGAPRAGDFR GTGHLTTTTT TQQNGGRAPF CESCKQQIRG AFVLATGKSW CPEHFVCANS 540
541 SCRRRLLECG FVEEDGQKFC ESCFEQHIAP RCNKCSKPII SDCLNALQKK WHPTCFTCAH 600
601 CQKPFGNSAF YLEQGLPYCE QDWNALFTTK CVSCRYPIEA GDRWVEALGN AFHSNCFTCA 660
661 RCNHNLEGES FFAKNGQPFC RLHA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
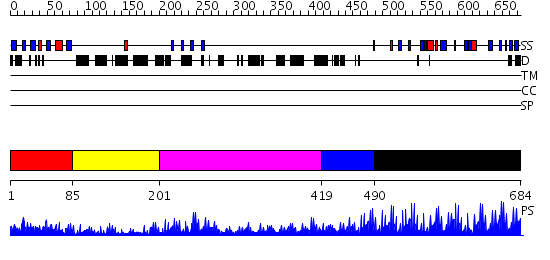
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..84] | 15.522879 | No description for 2uzcA was found. | |
| 2 | View Details | [85..200] | 3.22 | Cysteine-rich (intestinal) protein, CRP, CRIP | |
| 3 | View Details | [201..418] | 3.3 | No description for 2dfyX was found. | |
| 4 | View Details | [419..489] | 11.30103 | Five-finger GLI1 | |
| 5 | View Details | [490..684] | 43.09691 | No description for 2i13A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)