













| General Information: |
|
| Name(s) found: |
daf-16 /
CE38723
[WormBase]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 568 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
response to heat
[IEP dauer larval development [IGI insulin receptor signaling pathway [IDA determination of adult lifespan [IGI regulation of transcription, DNA-dependent [IEA positive regulation of transcription from RNA polymerase II promoter [IDA |
| Molecular Function: |
transcription factor activity
[IDA protein binding [IPI sequence-specific DNA binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQAWNCREVW RRIRKHLAHT SRTTVYSQFK FQIFEIFFQS FFPIFLHFAA KNSCFEPDFL 60
61 DSPLPSDITL HNLEPARPDS GMSFSTDFDD DFFNLDLHQQ ERSASFGGVT QYSQQFLREK 120
121 CSFSPYFHTS LETVDSGRTS LYGSNEQCGQ LGGASSNGST AMLHTPDGSN SHQTSFPSDF 180
181 RMSESPDDTV SGKKTTTRRN AWGNMSYAEL ITTAIMASPE KRLTLAQVYE WMVQNVPYFR 240
241 DKGDSNSSAG WKNSIRHNLS LHSRFMRIQN EGAGKSSWWV INPDAKPGRN PRRTRERSNT 300
301 IETTTKAQLE KSRRGAKKRI KERALMGSLH STLNGNSIAG SIQTISHDLY DDDSMQGAFD 360
361 NVPSSFRPRT QSNLSIPGSS SRVSPAIGSD IYDDLEFPSW VGESVPAIPS DIVDRTDQMR 420
421 IDATTHIGGV QIKQESKPIK TEPIAPPPSY HELNSVRGSC AQNPLLRNPI VPSTNFKPMP 480
481 LPGAYGNYQN GGITPINWLS TSNSSPLPGI QSCGIVAAQH TVASSSALPI DLENLTLPDQ 540
541 PLMDTMDVDA LIRHELSQAG GQHIHFDL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
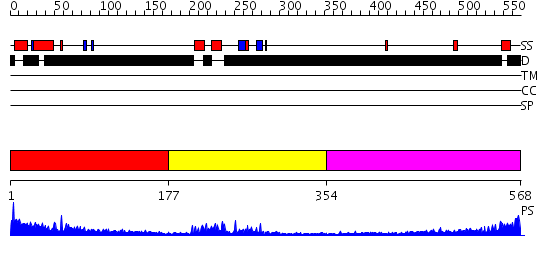
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..176] | 2.293997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [177..353] | 23.39794 | Afx (Foxo4) | |
| 3 | View Details | [354..568] | 6.417997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
|||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.79 |
Source: Reynolds et al. (2008)