













| General Information: |
|
| Name(s) found: |
hif-1 /
CE38181
[WormBase]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 665 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
response to hypoxia
[IMP signal transduction [IEA regulation of transcription, DNA-dependent [IEA regulation of transcription [IEA |
| Molecular Function: |
transcription factor activity
[ISS transcription regulator activity [IEA signal transducer activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEDNRKRNME RRRETSRHAA RDRRSKESDI FDDLKMCVPI VEEGTVTHLD RIALLRVAAT 60
61 ICRLRKTAGN VLENNLDNEI TNEVWTEDTI AECLDGFVMI VDSDSSILYV TESVAMYLGL 120
121 TQTDLTGRAL RDFLHPSDYD EFDKQSKMLH KPRGEDTDTT GINMVLRMKT VISPRGRCLN 180
181 LKSALYKSVS FLVHSKVSTG GHVSFMQGIT IPAGQGTTNA NASAMTKYTE SPMGAFTTRH 240
241 TCDMRITFVS DKFNYILKSE LKTLMGTSFY ELVHPADMMI VSKSMKELFA KGHIRTPYYR 300
301 LIAANDTLAW IQTEATTITH TTKGQKGQYV ICVHYVLGIQ GAEESLVVCT DSMPAGMQVD 360
361 IKKEVDDTRD YIGRQPEIVE CVDFTPLIEP EDPFDTVIEP VVGGEEPVKQ ADMGIYSVTD 420
421 EYEQRVYGQI SAGNDIEDCD MDSDTESIED SGLAPEFDVD LAPIYNEKME VDGEDEDGGE 480
481 EEHVRTATMT FCNGFFATSQ ALRRRHATGQ RTASGQPNRR ISVPLWPRRT SWIRHLVPPD 540
541 RRHHTDEEHK VREVEQLDRL PRQHRPHCHT VPTTRHLQRA SHNVDLTVPR VSNLDKSCTE 600
601 TLDLWDVAAI RPIRHDAFPP FHHQILSMCR PLEASTQSSE AMMCSPLCLL PILSPLPKGS 660
661 IRRQH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
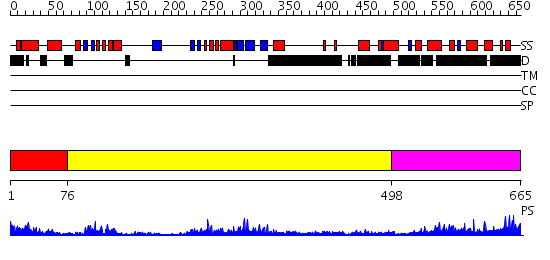
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..75] | 3.12 | Solution Structure of Max B-HLH-LZ | |
| 2 | View Details | [76..497] | 37.522879 | Crystal Structure of the PAS repeat region of the Drosophila clock protein PERIOD | |
| 3 | View Details | [498..665] | 14.69897 | No description for 2r6oA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)