













| General Information: |
|
| Name(s) found: |
nhr-23 /
CE37806
[WormBase]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 437 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
molting cycle, protein-based cuticle
[IMP locomotory behavior [IMP nematode larval development [IMP post-embryonic body morphogenesis [IMP regulation of transcription, DNA-dependent [IEA oviposition [IMP positive regulation of multicellular organism growth [IMP growth [IMP reproduction [IMP |
| Molecular Function: |
DNA binding
[IEA transcription factor activity [IEA zinc ion binding [IEA steroid hormone receptor activity [IEA sequence-specific DNA binding [IEA steroid binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRVSAQIEVI PCKVCGDKSS GVHYGVITCE GCKGFFRRSQ SSIVNYQCPR QKNCVVDRVN 60
61 RNRCQYCRLK KCIELGMSRD AVKFGRMSKK QREKVEDEVR MHKELAANGL GYQAIYGDYS 120
121 PPPSHPSYCF DQSMYGHYPS GTSTPVNGYS IAVAATPTTP MPQNMYGATP SSTNGTQYVA 180
181 HQATGGSFPS PQVPEEDVAT RVIRAFNQQH SSYTTQHGVC NVDPDCIPHL SRAGGWELFA 240
241 RELNPLIQAI IEFAKSIDGF MNLPQETQIQ LLKGSVFELS LVFAAMYYNV DAQAVCGERY 300
301 SVPFACLIAE DDAEMQLIVE VNNTLQEIVH LQPHQSELAL LAAGLILEQV SSSHGIGILD 360
361 TATIATAETL KNALYQSVMP RIGCMEDTIH RIQDVETRIR QTARLHQEAL QNFRMSDPTS 420
421 SEKLPALYKE LFTADRP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
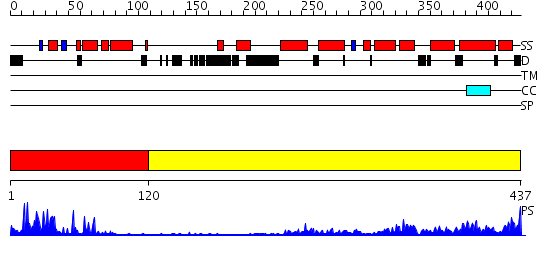
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..119] | 21.39794 | Human Liver Receptor Homologue DNA-Binding Domain (hLRH-1 DBD) in Complex with dsDNA from the hCYP7A1 Promoter | |
| 2 | View Details | [120..437] | 51.39794 | LIGAND-BINDING DOMAIN OF THE HUMAN NUCLEAR RECEPTOR RXR-ALPHA |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)