













| General Information: |
|
| Name(s) found: |
gi|481953
[NCBI NR]
gi|438212 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Plasmodium falciparum |
| Length: | 910 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
pentose-phosphate shunt
[TAS |
| Molecular Function: |
6-phosphogluconolactonase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDYENFVKSA EEINNLHNVN YLETKDLNDF NWKAAYYICK EIYDKQQINK DGYVVIGLSG 60
61 GRTPIDVYKN MCLIKDIKID KSKLIFFIID ERYKSDDHKF SNYNNIKFLF HNLNINEKEQ 120
121 LYKPDTTKSI VDCILDYNDK IKIMIEKYKK VDIAILGMGS DFHIASLFPN IFYNIYMNNY 180
181 QNNYIYNEKT LDFINNDQDN DNLKYLKEYV YFTTTNQFDV RKRITVSLNL LANASSKIFL 240
241 LNSKDKLDLW KNMLIKSYIE VNYNLYPATY LIDTSCTNEN VNINNNNNNN NKNKNNYCYS 300
301 NTTVISCGYE NYTKSIEEIY DSKYALSLYS NSLNKEELLT IIIFGCSGDL AKKKIYPALF 360
361 KLFCNNSLPK DLLIIGFART VQDFDTFFDK IVIYLKRCLL CYEDWSISKK KDLLNGFKNR 420
421 CRYFVGNYSS SESFENFNKY LTTIEEEEAK KKYYATCYKM NGSDYNISNN VAEDNISIDD 480
481 ENKTNEYFQM CTPKNCPDNV FSSNYNFPYV INRMLYLALP PHIFVSTLKN YKKNCLNSKG 540
541 TDKILLEKPF GNDLDSFKML SKQILENFNE QQIYRIDHYL GKDMVSGLLK LKFTNTFLLS 600
601 LMNRHFIKCI KITLKETKGV YGRGQYFDPY GIIRDVMQNH MLQLLTLITM EDPIDLNDES 660
661 VKNEKIKILK SIPSIKLEDT IIGQYEKAEN FKEDENNDDE SKKNHSYHDD PHIDKNSITP 720
721 TFCTCILYIN SINWYGVPII FKSGKGLNKD ICEIRIQFHN IMGSSDENMN NNEFVIILQP 780
781 VEAIYLKMMI KKTGCEEMEE VQLNLTVNEK NKKINVPEAY ETLLLECFKG HKKKFISDEE 840
841 LYESWRIFTP LLKELQEKQV KPLKYSFGSS GPKEVFGLVK KYYNYGKNYT HRPEFVRKSS 900
901 FYEDDLLDIN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
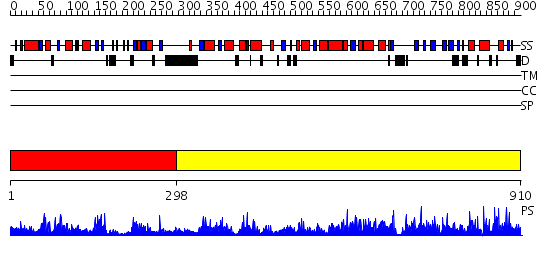
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..297] | 64.221849 | HUMAN GLUCOSAMINE-6-PHOSPHATE DEAMINASE ISOMERASE AT 1.75 A RESOLUTION COMPLEXED WITH N-ACETYL-GLUCOSAMINE-6-PHOSPHATE AND 2-DEOXY-2-AMINO-GLUCITOL-6-PHOSPHATE | |
| 2 | View Details | [298..910] | 1000.0 | Glucose 6-phosphate dehydrogenase, N-terminal domain; Glucose 6-phosphate dehydrogenase |
Functions predicted (by domain):
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)