













| General Information: |
|
| Name(s) found: |
AK2H_ECOLI
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Escherichia coli |
| Length: | 810 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSVIAQAGAK GRQLHKFGGS SLADVKCYLR VAGIMAEYSQ PDDMMVVSAA GSTTNQLINW 60
61 LKLSQTDRLS AHQVQQTLRR YQCDLISGLL PAEEADSLIS AFVSDLERLA ALLDSGINDA 120
121 VYAEVVGHGE VWSARLMSAV LNQQGLPAAW LDAREFLRAE RAAQPQVDEG LSYPLLQQLL 180
181 VQHPGKRLVV TGFISRNNAG ETVLLGRNGS DYSATQIGAL AGVSRVTIWS DVAGVYSADP 240
241 RKVKDACLLP LLRLDEASEL ARLAAPVLHA RTLQPVSGSE IDLQLRCSYT PDQGSTRIER 300
301 VLASGTGARI VTSHDDVCLI EFQVPASQDF KLAHKEIDQI LKRAQVRPLA VGVHNDRQLL 360
361 QFCYTSEVAD SALKILDEAG LPGELRLRQG LALVAMVGAG VTRNPLHCHR FWQQLKGQPV 420
421 EFTWQSDDGI SLVAVLRTGP TESLIQGLHQ SVFRAEKRIG LVLFGKGNIG SRWLELFARE 480
481 QSTLSARTGF EFVLAGVVDS RRSLLSYDGL DASRALAFFN DEAVEQDEES LFLWMRAHPY 540
541 DDLVVLDVTA SQQLADQYLD FASHGFHVIS ANKLAGASDS NKYRQIHDAF EKTGRHWLYN 600
601 ATVGAGLPIN HTVRDLIDSG DTILSISGIF SGTLSWLFLQ FDGSVPFTEL VDQAWQQGLT 660
661 EPDPRDDLSG KDVMRKLVIL AREAGYNIEP DQVRVESLVP AHCEGGSIDH FFENGDELNE 720
721 QMVQRLEAAR EMGLVLRYVA RFDANGKARV GVEAVREDHP LASLLPCDNV FAIESRWYRD 780
781 NPLVIRGPGA GRDVTAGAIQ SDINRLAQLL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
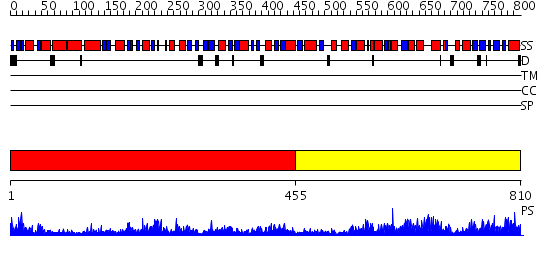
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..454] | 90.221849 | No description for 2hmfA was found. | |
| 2 | View Details | [455..810] | 68.522879 | Homoserine Dehydrogenase in complex with suicide inhibitor complex NAD-5-hydroxy-4-Oxonorvaline |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)