













| General Information: |
|
| Name(s) found: |
FBpp0298328
[FlyBase]
FBpp0298327 [FlyBase] |
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 730 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
compound eye morphogenesis
[IMP]
regulation of transcription [IDA][IEA] |
| Molecular Function: |
transcription regulator activity
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTESGIDLGF DMEFDLNINL LNDNDNMDFL PNVTENMEFY ELKSSSRCIR HNEIPTFKTA 60
61 TPTSRTQLKL QLQREQQQQM MIQQQTLDTA MDPKMHLLFG SGQGLMESEF IDSGSTSACG 120
121 SGSSSLEQMS QLVQMDNLID SSSGAKLKVP LQSIGVDVPP QVLQVSTVLE NPTRYHVIQK 180
181 QKNQVRQYLS ESFKPSMWGS HTSEIKLANN SASTGNLQNS SLQKGICDPL ERTNRFGCDS 240
241 AVSAKRIMPS DDAMPISPFG GSFVRCDDIN PIEPTVLRPN SHGAGEPENA HRTAQLGLSK 300
301 ANSSLSSTRS SSGIVNSIRI SSTSSSLQST SAPISPSVSS VATSVSELPS FDSDPDDIFD 360
361 DILQNDSFNF DKNFNSELSI KQEPQNLTDA EMNALAKDRQ KKDNHNMIER RRRFNINDRI 420
421 KELGTLLPKG SDAFYEVVRD IRPNKGTILK SSVDYIKCLK HEVTRLRQNE LRQRQVELQN 480
481 RKLMSRIKEL EMQAKSHGIL LSENHLTSLS APTQPYLKSF SLSPTASRSR RSLFDQPVEK 540
541 KIQVIDGADG NMGMNQVDEF MEDCKYAVQG GDPMLSSHSH MQSAPQSPSS KTLNSGSFEP 600
601 KNFSEADESL FRGKSSLASD DCCGINCSTS CYIQHQLTSR EGHPNHSHHT GIREIHSLSD 660
661 SAQNSEFSRL EHCDDPLLSS SHRSLGTVDE DQHNSVDMSA VMVHDSLSSL VDDNNSETMV 720
721 LASDTLDIEL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
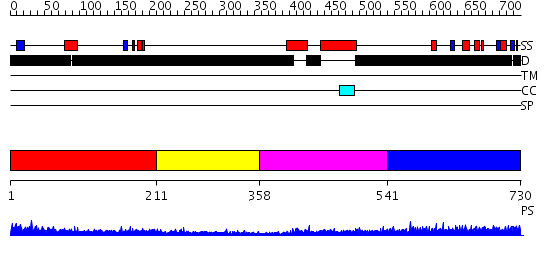
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..210] | 4.045757 | Sec24 | |
| 2 | View Details | [211..357] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [358..540] | 9.39794 | Solution Structure of Max B-HLH-LZ | |
| 4 | View Details | [541..730] | 5.127993 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)