













| General Information: |
|
| Name(s) found: |
acs-1 /
CE37772
[WormBase]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 578 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
propionate catabolic process, 2-methylcitrate cycle
[IEA teichoic acid biosynthetic process [IEA embryonic development ending in birth or egg hatching [IMP metabolic process [IEA menaquinone biosynthetic process [IEA siderophore biosynthetic process [IEA reproduction [IMP |
| Molecular Function: |
D-alanine-poly(phosphoribitol) ligase activity
[IEA AMP binding [IEA acetate-CoA ligase activity [IEA catalytic activity [IEA propionate-CoA ligase activity [IEA o-succinylbenzoate-CoA ligase activity [IEA (2,3-dihydroxybenzoyl)adenylate synthase activity [IEA ligase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVAPSRKSYV HGCSTVPLLF ETVGDRLRSA VDQVPDKEFL IFKREGIRKT YSQVATDAEN 60
61 LACGLLHLGL KKGDRIGIWG PNTYEWTTTQ FASALAGMVL VNINPSYQSE ELRYAIEKVG 120
121 IRALITPPGF KKSNYYQSIK DILPEVTLKE PGKSGITSRN FTCFQHLIMF DEEDKIYPGA 180
181 WKYTDVMKMG TEEDRHHLSK IERETQPDDS LNIQYTSGTT GQPKGATLTH HNVLNNAFFV 240
241 GLRAGYSEKK TIICIPNPLY HCFGCVMGVL AALTHLQTCV FPAPSFDALA ALQAIHEEKC 300
301 TALYGTPTMF IDMINHPEYA NYNYDSIRSG FIAGAPCPIT LCRRLVQDMH MTDMQVCYGT 360
361 TETSPVSFMS TRDDPPEQRI KSVGHIMDHL EAAIVDKRNC IVPRGVKGEV IVRGYSVMRC 420
421 YWNSEEQTKK EITQDRWYHT GDIAVMHDNG TISIVGRSKD MIVRGGENIY PTEVEQFLFK 480
481 HQSVEDVHIV GVPDERFGEV VCAWVRLHES AEGKTTEEDI KAWCKGKIAH FKIPRYILFK 540
541 KEYEFPLTVT GKVKKFEIRE MSKIELGLQQ VVSHFSEL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
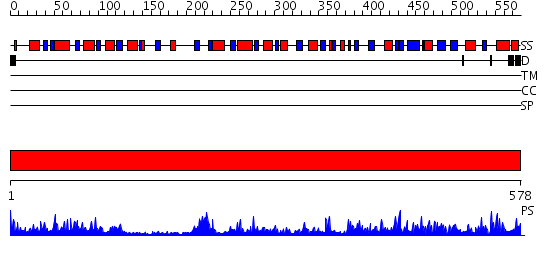
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..578] | 150.0 | No description for 2p2bA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.73 |
Source: Reynolds et al. (2008)