













| General Information: |
|
| Name(s) found: |
unc-62 /
CE37795
[WormBase]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 490 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
oocyte construction
[IMP locomotory behavior [IMP hermaphrodite genitalia development [IMP embryonic development ending in birth or egg hatching [IMP regulation of transcription, DNA-dependent [IEA oviposition [IMP cell migration [IMP positive regulation of multicellular organism growth [IMP |
| Molecular Function: |
transcription factor activity
[IEA sequence-specific DNA binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGGAPTSTPM MHHEMGEAMK RDKESIYAHP LYPLLVLLFE KCELATSTPR DTSRDGSTSS 60
61 DVCSSASFKD DLNEFVRHTQ ENADKQYYVP NPQLDQIMLQ SIQMLRFHLL ELEKVHELCD 120
121 NFCNRYVVCL KGKMPLDIVG DERASSSQPP MSPGSMGHLG HSSSPSMAGG ATPMHYPPPY 180
181 EPQSVPLPEN AGVMGGHPME GSSMAYSMAG MAAAAASSSS SSNQAGDHPL ANGGTLHSTA 240
241 GASQTLLPIA VSSPSTCSSG GLRQDSTPLS GETPMGHANG NSMDSISEAG DEFSVCGSND 300
301 DGRDSVLSDS ANGSQNGKRK VPKVFSKEAI TKFRAWLFHN LTHPYPSEEQ KKQLAKETGL 360
361 TILQVNNWFI NARRRIVQPM IDQNNRAGRS GQMNVCKNRR RNRSEQSPGP SPDSGSDSGA 420
421 NYSPDPSSLA ASTAMPYPAE FYMQRTMPYG GFPSFTNPAM PFMNPMMGFQ VAPTVDALSQ 480
481 QWVDLSAPHE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
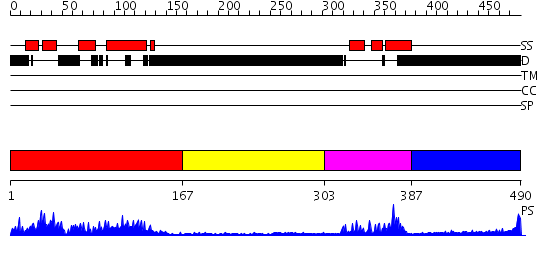
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..166] | 9.550979 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [167..302] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [303..386] | 19.69897 | Crystal Structure of HoxA9 and Pbx1 homeodomains bound to DNA | |
| 4 | View Details | [387..490] | 4.377996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)