













| General Information: |
|
| Name(s) found: |
gi|24374168
[NCBI NR]
gi|24348675 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 755 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
proteolysis
[ISS |
| Molecular Function: |
ATP binding
[ISS ATP-dependent peptidase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLNKDLEVTL NLAFQQAREA RHEYMTVEHL LLALIDNPAA QEALIACGAN LDKLRDEVAS 60
61 FIQQTTPIIA DMDDDKETQP TLGFQRVLQR AVFHVQSSGR NEVTGANVLV AIFSEQESQA 120
121 VYLLRRSDIS RLDVVNFISH GFSKNEDSDA HQDQERSEEQ AEASEERSML SQFATNLNQL 180
181 AKQGVIDPLI GRDQEIERAI QTLCRRRKNN PLLVGEAGVG KTAIAEGLAY RIVNHQVPEV 240
241 MAQATVYSLD LGSLLAGTKY RGDFEKRFKN LLKELSADKH AILFIDEIHT IIGAGAASGG 300
301 VMDASNLLKP LLSSGNLRCM GSTTFQEYQS IFEKDRALAR RFQKVDINEP SVAETTKILM 360
361 GLKSKYEEYH GVRYTQAAIN SAAVLSAKHI NDRHLPDKAI DVIDEAGARM AMLPQSKRKK 420
421 TIGQAEIETI VAKIARIPEK SVSATDKDLL RNLERNLKMV VFGQDKAIET LSSAIRLSRS 480
481 GLGTDKKPVG SFLFAGPTGV GKTEVTNQLA NCLGMKLVRF DMSEYMESHT VSRLIGAPPG 540
541 YVGYDQGGLL TDAVIKNPHC VVLLDEIEKA HPDVYNLLLQ VMDHGTLTDN NGRKADFRHV 600
601 TLVMTTNAGV QETIRKSIGF TQQDHTQDAL SEINRVFSPE FRNRLDSIIW FNHLDMTIIA 660
661 KVVDKFLVEL QAQLDAKHVV LEVSDEARTL LAQKGYDKSM GARPMARVVT ELIKRPLADE 720
721 LLFGELEHGG IAHVDAKDGE IVIHAERNEK TTSKA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
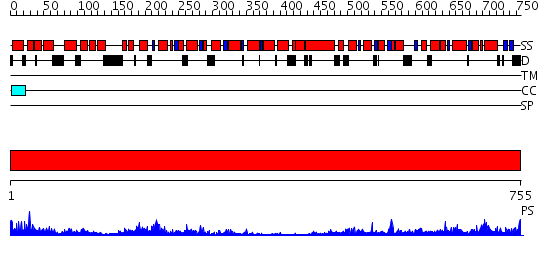
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..755] | 141.0 | N-terminal, ClpS-binding domain of ClpA, an Hsp100 chaperone; ClpA, an Hsp100 chaperone, AAA+ modules |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)