













| General Information: |
|
| Name(s) found: |
gi|24373705
[NCBI NR]
gi|24348070 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 712 amino acids |
Gene Ontology: |
|
| Cellular Component: |
exodeoxyribonuclease V complex
[ISS |
| Biological Process: |
DNA recombination
[ISS |
| Molecular Function: |
exodeoxyribonuclease V activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNNATALAAN WYNAGALRLK APLTELLKYW EKERLITALD RHFALEMARL HPNDAQEPLF 60
61 MLLCALLSQQ LSAQHTCLVL EYIVPTNPMA EPINRCQIRL SLTELIDALQ HFDAIGAPNS 120
121 NKPLIFEDGR LYLERYHHFE TQVASALTRL SAAALSLTPS QPLASDNQSL LQLRRQLDCL 180
181 FPSAAHTVEA NIDWQKVATA TALGNKLAVI TGGPGTGKTT TVTKLLLLQQ LGTPKQIRLV 240
241 APTGKAAARL SESIKASKAR LEQELQQASK ENPQENQVLL TALSRIPEEA STLHRLLGVI 300
301 PNSAHFRHHQ GNPLRLDLLI VDEASMVDLP MMYKLLSALP EHAGLILLGD QDQLASVEAG 360
361 AVLADICAGL KQASTQHTEW KMRYSQAQAE QLTALTGFNL SHFIHSEPKI GDSLCMLTHS 420
421 HRFKGDAGIG LLASAVNRAD LAGILSVWQS GLAELTWLEH SVDHSQTQAK ISEPSNNTGL 480
481 KALLAQACEQ YGDYLRLLQP THATPIHTSE PFCPPSAMEA IAKFNQYRIL CAMRTGDYGV 540
541 EGINLAVTKA LAQAKLIQPQ QEFYLGRPII IQSNDYNLGL FNGDIGLILQ DEERADRLMA 600
601 HFIKADGKLL KVLPARLPSH ETCYAMTVHK SQGSEFNRVA LVLPPAPSLV QWQLLTKELI 660
661 YTAITRAKQH FSCLGTQQVF ERASLQATQR ASGLADRLWG DQSLSHTAAV NS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
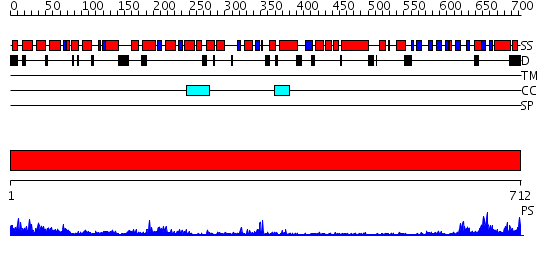
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..712] | 59.09691 | RecBCD:DNA complex |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)