













| General Information: |
|
| Name(s) found: |
GSHB_SHEON
[Swiss-Prot]
|
| Description(s) found:
Found 5 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 315 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
glutathione biosynthetic process
[ISS |
| Molecular Function: |
glutathione synthase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIKLGIVMDP ISEINIKKDS SFAMLMAAQE RGYQLFYMEM ADLAIVNGVA MGNMRPLKVM 60
61 NDANHWFELG EAKDTPLSEL NVVLMRKDPP FDTEYIYATY MLERAEEQGV LIVNKPQSLR 120
121 DANEKLFTAW FSEFTPETIV TRDANRIRAF HQAKGDIILK PLDGMGGTSI FRVKQDDPNL 180
181 GVIIETLTQY GNQYAMAQAF IPEITKGDKR ILVVDGEPVP YALARIPKKG ETRGNLAAGG 240
241 SGVAQPLSDS DWKIARAIGP ELKKRGLIFV GLDVIGDKLT EINVTSPTCI REIQAAFDVD 300
301 ITGMLFDAIE TRLGQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
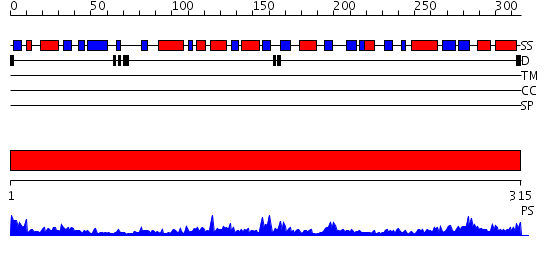
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..315] | 56.09691 | STRUCTURE OF GLUTATHIONE SYNTHETASE COMPLEXED WITH ADP AND GLUTATHIONE |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)