













| General Information: |
|
| Name(s) found: |
gi|24372020
[NCBI NR]
gi|24345884 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 677 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosolic pyruvate dehydrogenase complex
[ISS |
| Biological Process: |
acetyl-CoA biosynthetic process from pyruvate
[ISS |
| Molecular Function: |
dihydrolipoyllysine-residue acetyltransferase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAELKEVFVP DIGGDEVQVI EICAAVGDTL AAEESILTVE SDKATMDIPA PFAGVLAELK 60
61 VAVGDKVSEG TLIALIQAAG ASAQAAAPVA QAAAPAPAPV QAAPAPVVAA PATGATKLVE 120
121 AKVVEISVPD IGGDTDVSVI EVLVAAGDKI EVDAGLITLE TDKATMDVPS PFAGVVKEVK 180
181 VAVGDKVSQG SLVIMLEVGG AAPAAAPQAN APAASAPVAQ AAPAAAVAPV AAVPVVAVKE 240
241 IQVPDIGDAS NVDVIEVLVS VGDMISADQG LITLETDKAT MEVPAPFAGK LLSLTVKVGD 300
301 KVSQGSVIAT IETTSVATVS AGAATAPVAQ AAAPAPVAQE AAPAPVAAAP SRPPVPHHPS 360
361 AGAPVSTGAV HASPAVRRLA REFGVDLTQV TGSGRKGRIM KEDVQAYVKY ELSRPKATAA 420
421 TSVATGNGGG LQVIAAPKVD FSKFGEVEEI PLSRIQKISG PNLHRNWVTI PHVTQFDEAD 480
481 ITEMEEFRKQ QNDAAAKKKA DYKITPLVFM MKAVAKTLQQ FPVFNSSLSS DGESLIQKKY 540
541 FHIGVAVDTP NGLVVPVVRD VDKKGIIELS RELADISIRA RDGKLKSADM QGSCFTISSL 600
601 GGIGGTAFTP IVNYPDVAIL GVSKSEIKPK WNGKEFEPKL MLPLSLSYDH RVIDGAMAAR 660
661 FSVTLSGILS DIRTLIL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
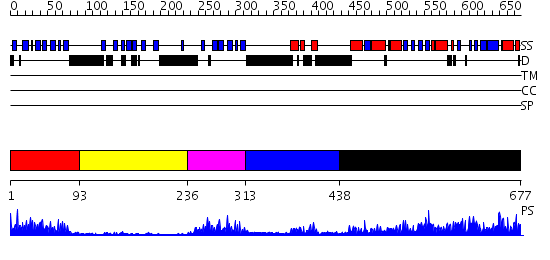
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..92] | 13.69897 | Ipoyl domain of dihydrolipoamide acetyltransferase | |
| 2 | View Details | [93..235] | 4.57 | The crystal structure of dihydrolipoamide dehydrogenase and dihydrolipoamide dehydrogenase-binding protein (didomain) subcomplex of human pyruvate dehydrogenase complex. | |
| 3 | View Details | [236..312] | 12.69897 | Biotin carboxyl carrier domain of transcarboxylase (TC 1.3S) | |
| 4 | View Details | [313..437] | 21.522879 | The crystal structure of dihydrolipoamide dehydrogenase and dihydrolipoamide dehydrogenase-binding protein (didomain) subcomplex of human pyruvate dehydrogenase complex. | |
| 5 | View Details | [438..677] | 73.39794 | CRYSTALLOGRAPHIC AND ENZYMATIC INVESTIGATIONS ON THE ROLE OF SER558, HIS610 AND ASN614 IN THE CATALYTIC MECHANISM OF AZOTOBACTER VINELANDII DIHYDROLIPOAMIDE ACETYLTRANSFERASE (E2P) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)