













| General Information: |
|
| Name(s) found: |
SET1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1080 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNYYRRAHA SSGSYRQPQE QPQYSRSGHY QYSNGHSHQQ YSSQYNQRRR YNHNDGTRRR 60
61 YNDDRPHSSN NASTRQYYAT NNSQSGPYVN KKSDISSRRG MSQSRYSNSN VHNTLASSSG 120
121 SLPTESALLL QQRPPSVLRY NTDNLKSKFH YFDPIKGEFF NKDKMLSWKA TDKEFSETGY 180
181 YVVKELQDGQ FKFKIKHRHP EIKASDPRNE NGIMTSGKVA THRKCRNSLI LLPRISYDRY 240
241 SLGPPPSCEI VVYPAQDSTT TNIQDISIKN YFKKYGEISH FEAFNDPNSA LPLHVYLIKY 300
301 ASSDGKINDA AKAAFSAVRK HESSGCFIMG FKFEVILNKH SILNNIISKF VEINVKKLQK 360
361 LQENLKKAKE KEAENEKAKE LQGKDITLPK EPKVDTLSHS SGSEKRIPYD LLGVVNNRPV 420
421 LHVSKIFVAK HRFCVEDFKY KLRGYRCAKF IDHPTGIYII FNDIAHAQTC SNAESGNLTI 480
481 MSRSRRIPIL IKFHLILPRF QNRTRFNKSS SSSNSTNVPI KYESKEEFIE ATAKQILKDL 540
541 EKTLHVDIKK RLIGPTVFDA LDHANFPELL AKRELKEKEK RQQIASKIAE DELKRKEEAK 600
601 RDFDLFGLYG GYAKSNKRNL KRHNSLALDH TSLKRKKLSN GIKPMAHLLN EETDSKETTP 660
661 LNDEGITRVS KEHDEEDENM TSSSSEEEEE EAPDKKFKSE SEPTTPESDH LHGIKPLVPD 720
721 QNGSSDVLDA SSMYKPTATE IPEPVYPPEE YDLKYSQTLS SMDLQNAIKD EEDMLILKQL 780
781 LSTYTPTVTP ETSAALEYKI WQSRRKVLEE EKASDWQIEL NGTLFDSELQ PGSSFKAEGF 840
841 RKIADKLKIN YLPHRRRVHQ PLNTVNIHNE RNEYTPELCQ REESSNKEPS DSVPQEVSSS 900
901 RDNRASNRRF QQDIEAQKAA IGTESELLSL NQLNKRKKPV MFARSAIHNW GLYALDSIAA 960
961 KEMIIEYVGE RIRQPVAEMR EKRYLKNGIG SSYLFRVDEN TVIDATKKGG IARFINHCCD1020
1021 PNCTAKIIKV GGRRRIVIYA LRDIAASEEL TYDYKFEREK DDEERLPCLC GAPNCKGFLN1080
1081 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
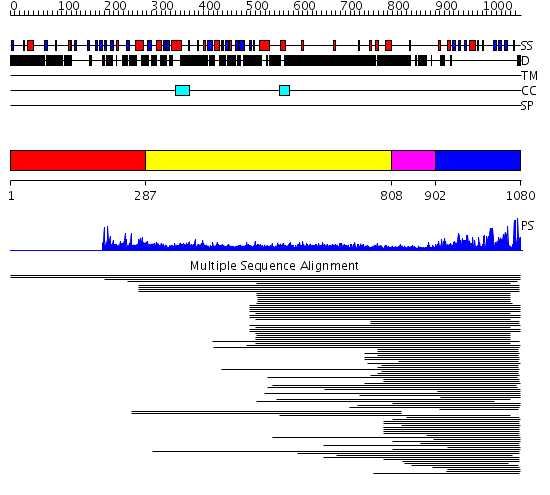
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..286] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [287..807] | 22.272997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [808..901] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [902..1080] | 62.69897 | SET domain No confident structure predictions are available. | |
| 5 | View Details | [734..809] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [810..1080] | 61.69897 | No description for 2r3aA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)