













| General Information: |
|
| Name(s) found: |
RUP1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 671 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMDNQAVKSL LEMGIPHEVA VDALQRTGGN LEAAVNFIFS NELPEQAEMG EENDGSQPRI 60
61 SENKIVAGTK PCDVPNNGDQ DIDMPDVSGV DVDYDDDEDI TDERSGSNST SGCRVTAQNY 120
121 DRYSISETSI PPPSYSIVQH NEFKSNVGDP TVVLPLPLNS LIESYFGLFA LLTAVYFPHV 180
181 FLKPDFKDLN YRADWFKGSS FTEPKYRLAY CEAEDGSTTS EIVLASGPNE GLQPHLLWQL 240
241 QKLISVVNTR KCERAFVSAK VFTSSLEPQL RSKLADSEHL YEVLPAFIKS LAVDLEMCPG 300
301 IRDRETRSLF ISSALHTPNK NEPPMETFLS LFHFLPEEYD SNLYKMFNVL LYPEEEEEEE 360
361 DVIRGGEQEE ARYVEPENTL KEVAPVLTIL FNELETNTES VSLPNGVDIP LEFYPQLYTK 420
421 QCKDQLIRHI ISKRKQARTR SRCLLQEINE LKSYQGKNIS TILESTLAYL QTIPDDANNE 480
481 AAKQIASLKD TLNSARAAKM EEYKDLASKL HGEWNLSHPE THIINTAKQL GLIENPYILT 540
541 MAALSPYSYF IRSRNGAWSW IQSNTLGTEF KVKKCSSPSV VQEAIKHGTK YASETPLMFI 600
601 YCEEGKIPTE EVVAEALKSN SGCLKFAEDD QNSLKTLRSQ FFDGMGDPEQ ATNNINNGND 660
661 NDNDDDIDSD N |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
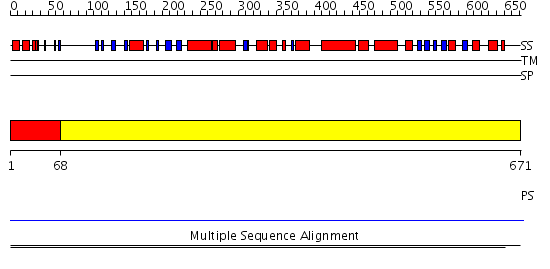
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..67] | 7.619789 | UBA/TS-N domain No confident structure predictions are available. | |
| 2 | View Details | [68..671] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [149..671] | 3.1 | Ubiquitin carboxyl-terminal hydrolase 7 (USP7, HAUSP) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)