













| General Information: |
|
| Name(s) found: |
MKT1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 830 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAIKSLESFL FERGLVGSYA IEALNNCTLD IDVNHYVSRL LTNKREQYLD AIGGFPTSLK 60
61 MYLESDLKIF KDFNITPIFV FNGGLTYNQL EASGHFTAAS ASASISSTTT SSSGTNATTR 120
121 SNTESVLLQR SRGWTQWNNL ISSNQNSYID QPIQPQEPFR HNTTIDSKAY QNDLIAYFIE 180
181 HGYMYQVAPY SSWFQLAYLL NSAYIDAIYG PTDCLMLDCV DRFILGMEFP NKEFRFIDRS 240
241 RVMKDLGCTH EEFIDIAMAV GNDLQPTTLP PLQIYPVPQL FDIALEMVLN TGTNFYAYQL 300
301 STTLQNDSKE NIQNYQRGIS ALRYMPVLKD TGKVELFVQE IVVSEEDSEK NNKDGKKSNL 360
361 SSPSSASSSA SPATTVTKNA SEKLTYEKSS TKEVRKPRDI PNDVHDFIGQ MLPHEYYFYR 420
421 SIGLVTGKLF DAIVTGVYPE EPPLGGGSST SYKKLVSKSV EIFKNKEINL LTQPINRYYQ 480
481 IKQIKQVKWY AANEPTTLTN RMSPSMFETI NHLIVKTETS DEKEFSISEF ITTINGSSNM 540
541 AKDFISEKVI FPNSVPIESK LNSPFNLLST NFLRLLVLLE FFTFDFKEKL LEPTRWGEVF 600
601 LKLNELNIDS KYHESVIIFL VFLKCDVLKL DEEVQPPAPS ALSQATLRSY PEESLYVLLI 660
661 TRVLTLFQVD QKPSNYHGPI DKKTLIFRDH LSFIKENLNE LFEAVLISSL TSGEFNRLSL 720
721 DNFGWARKIV RYLPFKLDSP NTIMAMMWEF FLQKYLHNGN AKNDALSLVA TEFNTYKSTP 780
781 NLDEQFVESH RFLLEISKVM QELNAAKLID ENVFKLFTKA VEFTTTALSS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
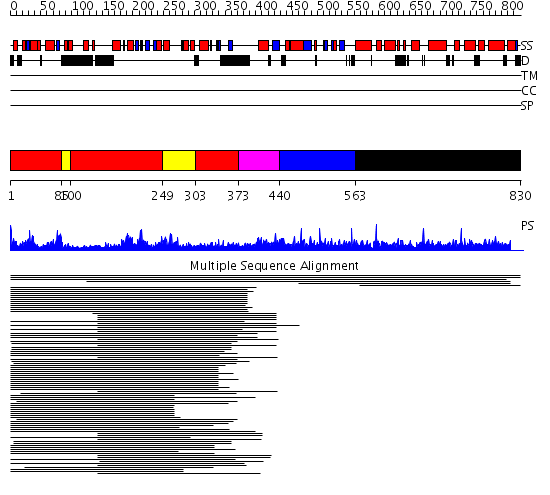
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..84] [100..248] [303..372] |
56.30103 | Flap endonuclease-1 (Fen-1 nuclease) | |
| 2 | View Details | [85..99] [249..302] |
56.30103 | Flap endonuclease-1 (Fen-1 nuclease) | |
| 3 | View Details | [373..439] | 12.522879 | Exonuclease domain of prokaryotic DNA polymerase; DNA polymerase I (Klenow fragment) | |
| 4 | View Details | [440..562] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [563..830] | 1.005883 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)