













| General Information: |
|
| Name(s) found: |
MBP1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 833 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNQIYSARY SGVDVYEFIH STGSIMKRKK DDWVNATHIL KAANFAKAKR TRILEKEVLK 60
61 ETHEKVQGGF GKYQGTWVPL NIAKQLAEKF SVYDQLKPLF DFTQTDGSAS PPPAPKHHHA 120
121 SKVDRKKAIR SASTSAIMET KRNNKKAEEN QFQSSKILGN PTAAPRKRGR PVGSTRGSRR 180
181 KLGVNLQRSQ SDMGFPRPAI PNSSISTTQL PSIRSTMGPQ SPTLGILEEE RHDSRQQQPQ 240
241 QNNSAQFKEI DLEDGLSSDV EPSQQLQQVF NQNTGFVPQQ QSSLIQTQQT ESMATSVSSS 300
301 PSLPTSPGDF ADSNPFEERF PGGGTSPIIS MIPRYPVTSR PQTSDINDKV NKYLSKLVDY 360
361 FISNEMKSNK SLPQVLLHPP PHSAPYIDAP IDPELHTAFH WACSMGNLPI AEALYEAGTS 420
421 IRSTNSQGQT PLMRSSLFHN SYTRRTFPRI FQLLHETVFD IDSQSQTVIH HIVKRKSTTP 480
481 SAVYYLDVVL SKIKDFSPQY RIELLLNTQD KNGDTALHIA SKNGDVVFFN TLVKMGALTT 540
541 ISNKEGLTAN EIMNQQYEQM MIQNGTNQHV NSSNTDLNIH VNTNNIETKN DVNSMVIMSP 600
601 VSPSDYITYP SQIATNISRN IPNVVNSMKQ MASIYNDLHE QHDNEIKSLQ KTLKSISKTK 660
661 IQVSLKTLEV LKESSKDENG EAQTNDDFEI LSRLQEQNTK KLRKRLIRYK RLIKQKLEYR 720
721 QTVLLNKLIE DETQATTNNT VEKDNNTLER LELAQELTML QLQRKNKLSS LVKKFEDNAK 780
781 IHKYRRIIRE GTEMNIEEVD SSLDVILQTL IANNNKNKGA EQIITISNAN SHA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
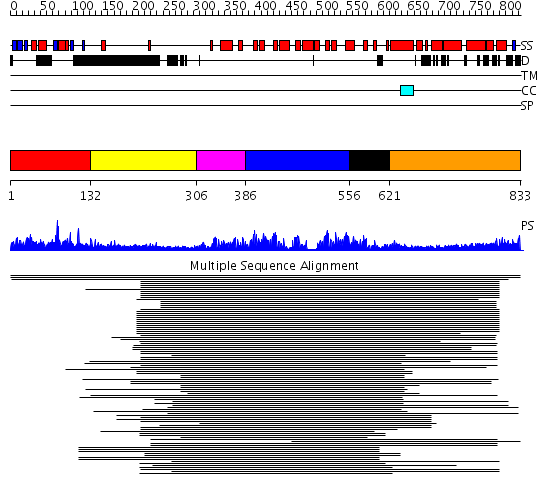
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..131] | 725.67837 | DNA-binding domain of Mlu1-box binding protein MBP1 | |
| 2 | View Details | [132..305] | 1.274996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [306..385] | 21.69897 | p18ink4C(ink6) | |
| 4 | View Details | [386..555] | 159.38764 | Swi6 ankyrin-repeat fragment | |
| 5 | View Details | [556..620] | 13.0 | Pyk2-associated protein beta; Pyk2-associated protein beta ARF-GAP domain | |
| 6 | View Details | [621..833] | 2.342996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)