













| General Information: |
|
| Name(s) found: |
IXR1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 597 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNTGISPKQD DASNSNLLNI GQDHSLQYQG LEHNDSQYRD ASHQTPHQYL NQFQAQPQQQ 60
61 QQQQQQQQQQ QQQAPYQGHF QQSPQQQQQN VYYPLPPQSL TQPTSQSQQQ QQQQQQQQYA 120
121 NSNSNSNNNV NVNALPQDFG YMQQTGSGQN YPTINQQQFS EFYNSFLSHL TQKQTNPSVT 180
181 GTGASSNNNS NNNNVSSGNN STSSNPAQLA ASQLNPATAA AAAANNAAGP ASYLSQLPQV 240
241 QRYYPNNMNA LSSLLDPSSA GNAAGNANTA THPGLLPPNL QPQLTHHQQQ MQQQLQLQQQ 300
301 QQLQQQQQLQ QQHQLQQQQQ LQQQHHHLQQ QQQQQQHPVV KKLSSTQSRI ERRKQLKKQG 360
361 PKRPSSAYFL FSMSIRNELL QQFPEAKVPE LSKLASARWK ELTDDQKKPF YEEFRTNWEK 420
421 YRVVRDAYEK TLPPKRPSGP FIQFTQEIRP TVVKENPDKG LIEITKIIGE RWRELDPAKK 480
481 AEYTETYKKR LKEWESCYPD ENDPNGNPTG HSHKAMNMNL NMDTKIMENQ DSIEHITANA 540
541 IDSVTGSNSN STNPNTPVSP PISLQQQPLQ QQQQQQQQQQ HMLLADPTTN GSIIKNE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
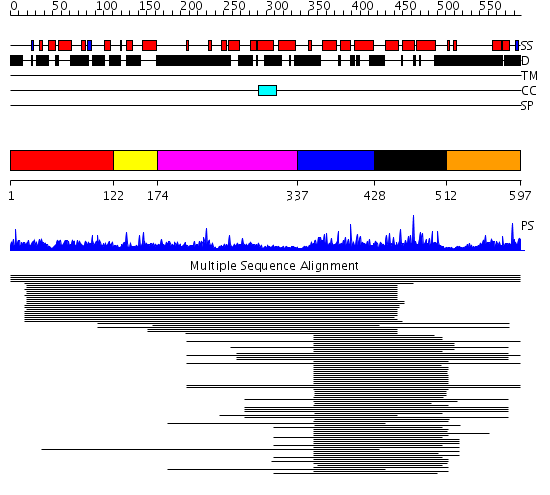
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..121] | 1.017991 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [122..173] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [174..336] | 39.071898 | Upstream binding factor | |
| 4 | View Details | [337..427] | 58.045757 | Sox-5 | |
| 5 | View Details | [428..511] | 41.29073 | NHP6a | |
| 6 | View Details | [512..597] | 1.177993 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)