













| General Information: |
|
| Name(s) found: |
CEX1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 761 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNFSSIFKSI SNFQFPYTIE ETAITETALW QCFDGTRKAD SLPVTVFKAK RSPENESLIL 60
61 NAVHKSKILK IPGLCTVLET FDSDPQSTFI VTERVVPFPW DNLGSLSQNK FGVELGISQL 120
121 LATLGFLKNF VLGTLSKDSV FINIKGEWVL FGLELCSSKE GLSAFEFASR ARSYYNIIGS 180
181 QLPCEDPNTI DSMGLGLLIK SLMAPSCLPK DWIVNVNMIS DGKITIENFR KRLENTETWR 240
241 SNPLINFYQE LRELHIKDPQ GKLVVMSNLE NLYLESREIF RNLTPGMIEN FIIPELCEII 300
301 KLLMTQSISS AASPIGMNFN ASHKLVPFLA IVLDLTSETN TFPVGFNDLI TQSFKLPDRQ 360
361 VRFLLLIYLP KLIGPLSKSE ISSRIYPHFI QGLTDSDATL RLQTLKTIPC IVSCLTERQL 420
421 NNELLRFLAK TQVDSDVEIR TWTVIIISKI STILSTSVGN RSNILATAFT KSLKDPQVKP 480
481 RLAALYGLEK SIELFDVNTI ANKILTVIAP GLLDKSPIVR GRAKILFEEY LEKLEKEAQL 540
541 IQTNDSTADS EDVKDIDFEN YGCDEEDMNK EDNLLAAQFL NNLRLNSPSA TTPSNITESE 600
601 IDSAQDGSGW DDLSDTDGFI TNGTTESFDE TTNPVTTAST PKLFGKPIKI NKSWNDELND 660
661 DGWIQDESGP SKVPQKHTRP QNSTLAKSIA PSSRLSIKKK KTTILAPRNI ASNSTVTTKS 720
721 SLSNKTARSK PISSIRGSVT KKGNVDGWDD DGDSDSWDTN W |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
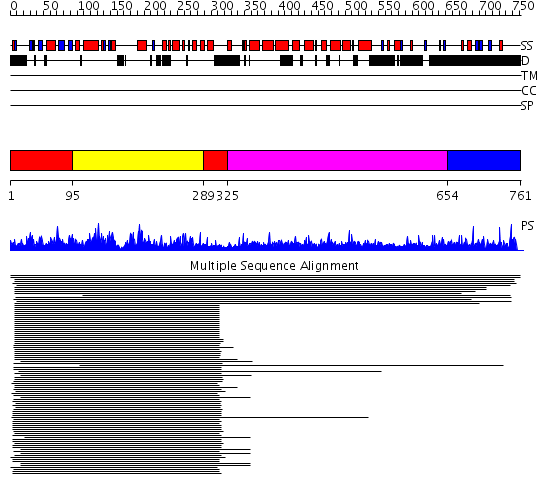
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..94] [289..324] |
105.0 | MAP kinase p38 | |
| 2 | View Details | [95..288] | 105.0 | MAP kinase p38 | |
| 3 | View Details | [325..653] | 7.221849 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 4 | View Details | [654..761] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)