













| General Information: |
|
| Name(s) found: |
BAS1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 811 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNISTKDIR KSKPKRGSGF DLLEVTESLG YQTHRKNGRN SWSKDDDNML RSLVNESAKE 60
61 LGYENGLEDV KTIQQSNHLS KCIAWDVLAT RFKHTVRTSK DVRKRWTGSL DPNLKKGKWT 120
121 QEEDEQLLKA YEEHGPHWLS ISMDIPGRTE DQCAKRYIEV LGPGSKGRLR EWTLEEDLNL 180
181 ISKVKAYGTK WRKISSEMEF RPSLTCRNRW RKIITMVVRG QASEVITKAI KENKNIDMTD 240
241 GKLRQHPIAD SDIRSDSTPN KEEQLQLSQQ NNPSLIKQDI LNVKENESSK LPRLKDNDGP 300
301 ILNDSKPQAL PPLKEISAPP PIRMTQVGQT HTSGSIRSKV SLPIEGLSQM NKQSPGGISD 360
361 SPQTSLPPAF NPASLDEHMM NSNSISDSPK HAYSTVKTRE PNSSSTQWKF TLKDGQGLSI 420
421 SNGTIDSTKL VKELVDQAKK YSLKISIHQH IHNHYVTSTD HPVSSNTGLS NIGNINGNPL 480
481 LMDSFPHMGR QLGNGLPGLN SNSDTFNPEY RTSLDNMDSD FLSRTPNYNA FSLEATSHNP 540
541 ADNANELGSQ SNRETNSPSV FYPQANTLIP TNSTATNNEI IQGNVSANSM SPNFNGTNGK 600
601 APSSTASYTT SGSEMPPDVG PNRIAHFNYL PPTIRPHLGS SDATRGADLN KLLNPSPNSV 660
661 RSNGSKTKKK EKRKSESSQH HSSSSVTTNK FNHIDQSEIS RTTSRSDTPL RDEDGLDFWE 720
721 TLRSLATTNP NPPVEKSAEN DGAKPQVVHQ GIGSHTEDSS LGSHSGGYDF FNELLDKKAD 780
781 TLHNEAKKTS EHDMTSGGST DNGSVLPLNP S |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
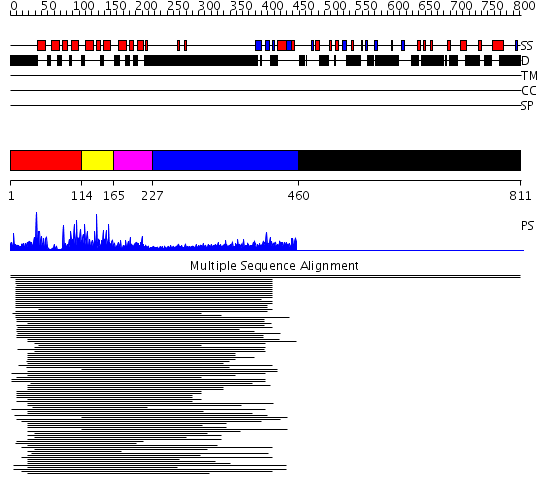
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..113] | 202.751666 | c-Myb, DNA-binding domain | |
| 2 | View Details | [114..164] | 202.751666 | c-Myb, DNA-binding domain | |
| 3 | View Details | [165..226] | 202.751666 | c-Myb, DNA-binding domain | |
| 4 | View Details | [227..459] | 3.220997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [460..811] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)