













| General Information: |
|
| Name(s) found: |
APC5_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 685 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKYGPLGIT NFITPYDLCI LILIHAHCSQ DNGISVPTAV FLRLISPTRP SLEWNPLLKD 60
61 NSNLRSSSIV PPPVLPILDN IIRILLDDKD GNKIALTLMG YLEAINGLDS INRLMMDLEK 120
121 NCLVNNYRSM KMRTTSTRRQ MTRASFLGTF LSTCIRKYQI GDFEMRETIW INLQNFKTVF 180
181 KHTPLWLRFK DNVHIQKVKN CLLANDEISV EDQQMVEFFQ HFNNGNDADS KTMNEENYGT 240
241 LISIQHLQSI VNRQIVNWLD NTEFNLMGQE ETSSTYEEQS GLVFDLLDTL SLNDATKFPL 300
301 IFILKYLEAI KENSYQTALD SLHNYFDYKS TGNSQNYFHI SLLSLATFHS SFNECDAAIN 360
361 SFEEATRIAR ENKDMETLNL IMIWIINFIE VHPEYANRFY ITVEQIIKYL KNSSDVEDAN 420
421 IFSNAYKFET LLSMVKESKT AEVSSSLLKF MAITLQNVPS QNFDLFQSLV SYEVKFWKEL 480
481 GYESISDVYE KFLSKTSSSS LRNYDSSIIN QDIKVAFKAL EEDDFLKVKQ YLLKSESLEL 540
541 DYDQKINLKY LRVKYLVKIG DYDLSMRLIN QYVKECCEEV ADSNWRFKFE IESINVLLLS 600
601 DVGIRSLPKI IKLIDEYKEI GNPLRCVILL LKLCEVLIQV GKSMEAECLI SCNLSTILEF 660
661 PFVRKKTDEL LESLSVEEDR DVQMT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
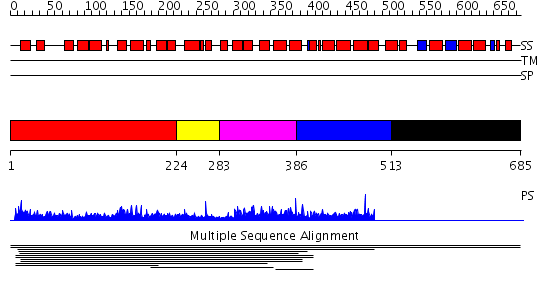
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..223] | 1.00996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [224..282] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [283..385] | 9.5 | Transcription factor MalT domain III | |
| 4 | View Details | [386..512] | 9.5 | Transcription factor MalT domain III | |
| 5 | View Details | [513..685] | 9.5 | Transcription factor MalT domain III |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.75 |
Source: Reynolds et al. (2008)