













| General Information: |
|
| Name(s) found: |
GCN5_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 568 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
histone acetyltransferase complex [IPI] |
| Biological Process: |
flower development
[IMP]
positive regulation of transcription [TAS] histone acetylation [IDA] response to light stimulus [IMP] root morphogenesis [IMP] |
| Molecular Function: |
histone acetyltransferase activity
[IDA][ISS]
DNA binding [IDA] H3 histone acetyltransferase activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSHSSHLNA ANRSRSSQTP SPSHSASASV TSSLHKRKLA ATTAANAAAS EDHAPPSSSF 60
61 PPSSFSADTR DGALTSNDEL ESISARGADT DSDPDESEDI VVDDDEDEFA PEQDQDSSIR 120
121 TFTAARLDSS SGVNGSSRNT KLKTESSTVK LESSDGGKDG GSSVVGTGVS GTVGGSSISG 180
181 LVPKDESVKV LAENFQTSGA YIAREEALKR EEQAGRLKFV CYSNDSIDEH MMCLIGLKNI 240
241 FARQLPNMPK EYIVRLLMDR KHKSVMVLRG NLVVGGITYR PYHSQKFGEI AFCAITADEQ 300
301 VKGYGTRLMN HLKQHARDVD GLTHFLTYAD NNAVGYFVKQ GFTKEIYLEK DVWHGFIKDY 360
361 DGGLLMECKI DPKLPYTDLS SMIRQQRKAI DERIRELSNC QNVYPKIEFL KNEAGIPRKI 420
421 IKVEEIRGLR EAGWTPDQWG HTRFKLFNGS ADMVTNQKQL NALMRALLKT MQDHADAWPF 480
481 KEPVDSRDVP DYYDIIKDPI DLKVIAKRVE SEQYYVTLDM FVADARRMFN NCRTYNSPDT 540
541 IYYKCATRLE THFHSKVQAG LQSGAKSQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
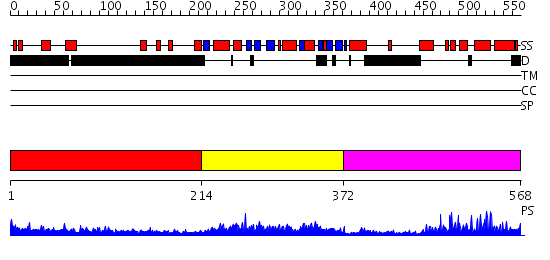
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..213] | 16.150997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [214..371] | 75.221849 | Catalytic domain of GCN5 histone acetyltransferase | |
| 3 | View Details | [372..568] | 30.39794 | TAFII250 double bromodomain module |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)