













| General Information: |
|
| Name(s) found: |
MSH1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1118 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
mitochondrion [IDA] |
| Biological Process: |
mitochondrial DNA metabolic process
[IMP]
mismatch repair [ISS] mitochondrial genome maintenance [IMP] |
| Molecular Function: |
ATP binding
[IEA][ISS]
catalytic activity [IEA] nuclease activity [IEA] mismatched DNA binding [IEA] damaged DNA binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHWIATRNAV VSFPKWRFFF RSSYRTYSSL KPSSPILLNR RYSEGISCLR DGKSLKRITT 60
61 ASKKVKTSSD VLTDKDLSHL VWWKERLQTC KKPSTLQLIE RLMYTNLLGL DPSLRNGSLK 120
121 DGNLNWEMLQ FKSRFPREVL LCRVGEFYEA IGIDACILVE YAGLNPFGGL RSDSIPKAGC 180
181 PIMNLRQTLD DLTRNGYSVC IVEEVQGPTP ARSRKGRFIS GHAHPGSPYV YGLVGVDHDL 240
241 DFPDPMPVVG ISRSARGYCM ISIFETMKAY SLDDGLTEEA LVTKLRTRRC HHLFLHASLR 300
301 HNASGTCRWG EFGEGGLLWG ECSSRNFEWF EGDTLSELLS RVKDVYGLDD EVSFRNVNVP 360
361 SKNRPRPLHL GTATQIGALP TEGIPCLLKV LLPSTCSGLP SLYVRDLLLN PPAYDIALKI 420
421 QETCKLMSTV TCSIPEFTCV SSAKLVKLLE QREANYIEFC RIKNVLDDVL HMHRHAELVE 480
481 ILKLLMDPTW VATGLKIDFD TFVNECHWAS DTIGEMISLD ENESHQNVSK CDNVPNEFFY 540
541 DMESSWRGRV KGIHIEEEIT QVEKSAEALS LAVAEDFHPI ISRIKATTAS LGGPKGEIAY 600
601 AREHESVWFK GKRFTPSIWA GTAGEDQIKQ LKPALDSKGK KVGEEWFTTP KVEIALVRYH 660
661 EASENAKARV LELLRELSVK LQTKINVLVF ASMLLVISKA LFSHACEGRR RKWVFPTLVG 720
721 FSLDEGAKPL DGASRMKLTG LSPYWFDVSS GTAVHNTVDM QSLFLLTGPN GGGKSSLLRS 780
781 ICAAALLGIS GLMVPAESAC IPHFDSIMLH MKSYDSPVDG KSSFQVEMSE IRSIVSQATS 840
841 RSLVLIDEIC RGTETAKGTC IAGSVVESLD TSGCLGIVST HLHGIFSLPL TAKNITYKAM 900
901 GAENVEGQTK PTWKLTDGVC RESLAFETAK REGVPESVIQ RAEALYLSVY AKDASAEVVK 960
961 PDQIITSSNN DQQIQKPVSS ERSLEKDLAK AIVKICGKKM IEPEAIECLS IGARELPPPS1020
1021 TVGSSCVYVM RRPDKRLYIG QTDDLEGRIR AHRAKEGLQG SSFLYLMVQG KSMACQLETL1080
1081 LINQLHEQGY SLANLADGKH RNFGTSSSLS TSDVVSIL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
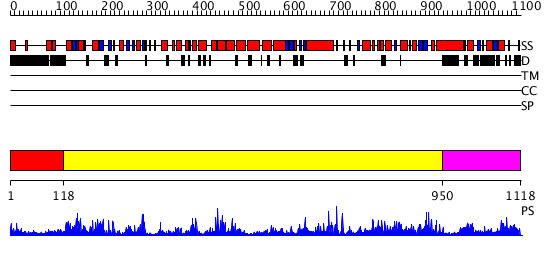
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..117] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [118..949] | 1000.0 | DNA repair protein MutS, domain III; DNA repair protein MutS, the C-terminal domain; DNA repair protein MutS, domain II; DNA repair protein MutS, domain I | |
| 3 | View Details | [950..1118] | 5.0 | Crystal Structure Analysis of ClpB |
| Source: Reynolds et al. 2008. Manuscript submitted |

|