













| General Information: |
|
| Name(s) found: |
DPO5_YEAST
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1022 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTGKVNRDLF FKLASDLREE RLHAAVALIK DLSALDLPDD AEEWSYVLNR LIKGLSSDRN 60
61 SARLGFSLCL TEVINLAVNM PPGQRPKGLE STNEFLSTLS TILNVNVNEG TKKSMKGKDE 120
121 RGILFGKLFG LKSLLNEPLF SEIFVKDLEK GNTEFFIRFT EQLIDLALKK NWIKEPCFFT 180
181 LFQTMKMLLP FMDESSAEKI LLIYDKYDLT LTNEGLSTYL LLKYEGDESL IPSVLDLKNP 240
241 GWKDNDPLAR GNLPLLTKVL RNSSVIPDAN GGLKETKKQK NTNWNPRLHF VWSVLLPLFG 300
301 NGKLENTSHI SKKRKKTNNK KVQNSIQFPE FWKMAVDESF FNEKASSERK YLGFLIIDAA 360
361 FKAVPGSYIG FCFSQNVMRT LINQSIDSQR VLNKISQLTL DSIVKACEED SANRLVPCLN 420
421 AMLFGPHGSI NFDKLTKSGT VSKLIAIKEL PSTVLAQLLD VFFLQLQDKK GVLSHTLFAL 480
481 DSILHIVRAH KVEINDMDIM KPVLRPIVYM AFFKHTSDDL KLEQLHELAK ERLYSILGEL 540
541 TINKEIRCKD PEINSWQYLT LKLILDIENS HVGDLINPLD ENLENIKNEA ISCLSKVCRS 600
601 RTAQSWGLST LLSMCLVQLY AGDTDSISVI EELCEFSKHE NNSMVGITEI LLSLLAQKKA 660
661 LLRKLSLIIW QQFIEEVGLE ELQILLDILK ARENKQGFAQ LFEGEEEFEE IKEENDASED 720
721 ESKTGSESES ESESDSDDAD EKDEEDEANE DILNIDKEAT SALVKALNLP DNIVNDKGEV 780
781 DLDQLEGLSD DGGDDEDEES MDDEKMMELD DQLSEIFKRR KEALSSISTG NQRKFEVKQS 840
841 RENVISFKHR VVDMLAVYVK YCEKLTLANK SEHSNNLGGS LSKLVYFIIP MLKCVNETLD 900
901 RPLADKISKL LKGKIFKIKV TAFKDMNKDI ELMDLLKKTH KLMLTSKPGQ HAAVFYSMCS 960
961 TSSLFLSKLY VEIGGNDKLD ELIDLYTATT KEWMQKGKCG PNIFIDFINW LSSKKQTVMD1020
1021 KE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
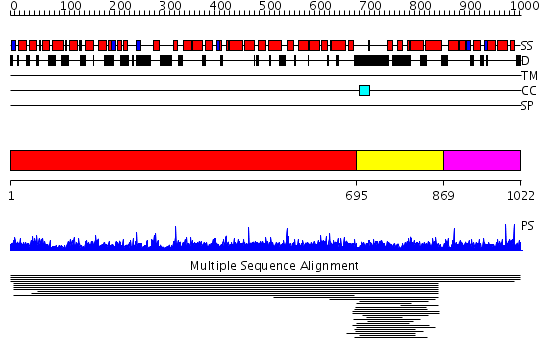
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..694] | 1.008835 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [695..868] | 5.016997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [869..1022] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [866..1022] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)