













| General Information: |
|
| Name(s) found: |
RAD25_YEAST
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 843 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTDVEGYQPK SKGKIFPDMG ESFFSSDEDS PATDAEIDEN YDDNRETSEG RGERDTGAMV 60
61 TGLKKPRKKT KSSRHTAADS SMNQMDAKDK ALLQDTNSDI PADFVPDSVS GMFRSHDFSY 120
121 LRLRPDHASR PLWISPSDGR IILESFSPLA EQAQDFLVTI AEPISRPSHI HEYKITAYSL 180
181 YAAVSVGLET DDIISVLDRL SKVPVAESII NFIKGATISY GKVKLVIKHN RYFVETTQAD 240
241 ILQMLLNDSV IGPLRIDSDH QVQPPEDVLQ QQLQQTAGKP ATNVNPNDVE AVFSAVIGGD 300
301 NEREEEDDDI DAVHSFEIAN ESVEVVKKRC QEIDYPVLEE YDFRNDHRNP DLDIDLKPST 360
361 QIRPYQEKSL SKMFGNGRAR SGIIVLPCGA GKTLVGITAA CTIKKSVIVL CTSSVSVMQW 420
421 RQQFLQWCTL QPENCAVFTS DNKEMFQTES GLVVSTYSMV ANTRNRSHDS QKVMDFLTGR 480
481 EWGFIILDEV HVVPAAMFRR VVSTIAAHAK LGLTATLVRE DDKIGDLNFL IGPKLYEANW 540
541 MELSQKGHIA NVQCAEVWCP MTAEFYQEYL RETARKRMLL YIMNPTKFQA CQFLIQYHER 600
601 RGDKIIVFSD NVYALQEYAL KMGKPFIYGS TPQQERMNIL QNFQYNDQIN TIFLSKVGDT 660
661 SIDLPEATCL IQISSHYGSR RQEAQRLGRI LRAKRRNDEG FNAFFYSLVS KDTQEMYYST 720
721 KRQAFLVDQG YAFKVITHLH GMENIPNLAY ASPRERRELL QEVLLKNEEA AGIEVGDDAD 780
781 NSVGRGSNGH KRFKSKAVRG EGSLSGLAGG EDMAYMEYST NKNKELKEHH PLIRKMYYKN 840
841 LKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
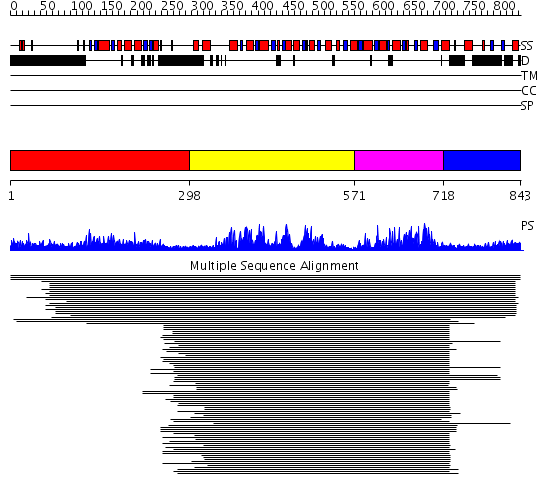
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..297] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [298..570] | 94.39794 | Putative DEAD box RNA helicase | |
| 3 | View Details | [571..717] | 94.39794 | Putative DEAD box RNA helicase | |
| 4 | View Details | [718..843] | 10.30103 | Nucleotide excision repair enzyme UvrB |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)