













| General Information: |
|
| Name(s) found: |
SGS1_YEAST
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1447 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVTKPSHNLR REHKWLKETA TLQEDKDFVF QAIQKHIANK RPKTNSPPTT PSKDECGPGT 60
61 TNFITSIPAS GPTNTATKQH EVMQTLSNDT EWLSYTATSN QYADVPMVDI PASTSVVSNP 120
121 RTPNGSKTHN FNTFRPHMAS SLVENDSSRN LGSRNNNKSV IDNSSIGKQL ENDIKLEVIR 180
181 LQGSLIMALK EQSKLLLQKC SIIESTSLSE DAKRLQLSRD IRPQLSNMSI RIDSLEKEII 240
241 KAKKDGMSKD QSKGRSQVSS QDDNIISSIL PSPLEYNTSS RNSNLTSTTA TTVTKALAIT 300
301 GAKQNITNNT GKNSNNDSNN DDLIQVLDDE DDIDCDPPVI LKEGAPHSPA FPHLHMTSEE 360
361 QDELTRRRNM RSREPVNYRI PDRDDPFDYV MGKSLRDDYP DVEREEDELT MEAEDDAHSS 420
421 YMTTRDEEKE ENELLNQSDF DFVVNDDLDP TQDTDYHDNM DVSANIQESS QEGDTRSTIT 480
481 LSQNKNVQVI LSSPTAQSVP SNGQNQIGVE HIDLLEDDLE KDAILDDSMS FSFGRQHMPM 540
541 SHSDLELIDS EKENEDFEED NNNNGIEYLS DSDLERFDEE RENRTQVADI QELDNDLKII 600
601 TERKLTGDNE HPPPSWSPKI KREKSSVSQK DEEDDFDDDF SLSDIVSKSN LSSKTNGPTY 660
661 PWSDEVLYRL HEVFKLPGFR PNQLEAVNAT LQGKDVFVLM PTGGGKSLCY QLPAVVKSGK 720
721 THGTTIVISP LISLMQDQVE HLLNKNIKAS MFSSRGTAEQ RRQTFNLFIN GLLDLVYISP 780
781 EMISASEQCK RAISRLYADG KLARIVVDEA HCVSNWGHDF RPDYKELKFF KREYPDIPMI 840
841 ALTATASEQV RMDIIHNLEL KEPVFLKQSF NRTNLYYEVN KKTKNTIFEI CDAVKSRFKN 900
901 QTGIIYCHSK KSCEQTSAQM QRNGIKCAYY HAGMEPDERL SVQKAWQADE IQVICATVAF 960
961 GMGIDKPDVR FVYHFTVPRT LEGYYQETGR AGRDGNYSYC ITYFSFRDIR TMQTMIQKDK1020
1021 NLDRENKEKH LNKLQQVMAY CDNVTDCRRK LVLSYFNEDF DSKLCHKNCD NCRNSANVIN1080
1081 EERDVTEPAK KIVKLVESIQ NERVTIIYCQ DVFKGSRSSK IVQANHDTLE EHGIGKSMQK1140
1141 SEIERIFFHL ITIRVLQEYS IMNNSGFASS YVKVGPNAKK LLTGKMEIKM QFTISAPNSR1200
1201 PSTSSSFQAN EDNIPVIAQK STTIGGNVAA NPPRFISAKE HLRSYTYGGS TMGSSHPITL1260
1261 KNTSDLRSTQ ELNNLRMTYE RLRELSLNLG NRMVPPVGNF MPDSILKKMA AILPMNDSAF1320
1321 ATLGTVEDKY RRRFKYFKAT IADLSKKRSS EDHEKYDTIL NDEFVNRAAA SSNGIAQSTG1380
1381 TKSKFFGANL NEAKENEQII NQIRQSQLPK NTTSSKSGTR SISKSSKKSA NGRRGFRNYR1440
1441 GHYRGRK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
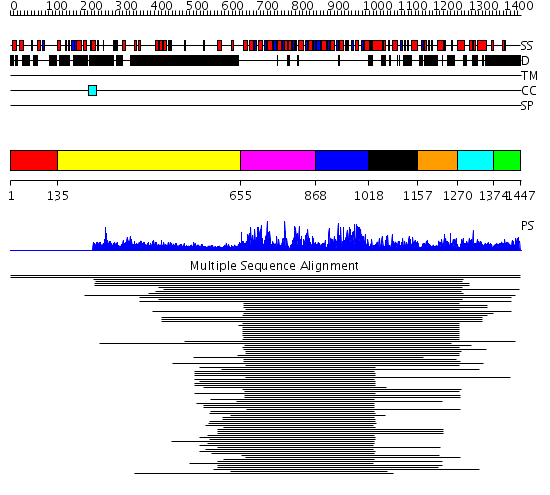
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..134] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [135..654] | 8.466997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [655..867] | 197.9691 | Putative DEAD box RNA helicase | |
| 4 | View Details | [868..1017] | 197.9691 | Putative DEAD box RNA helicase | |
| 5 | View Details | [1018..1156] | 14.69897 | Nucleotide excision repair enzyme UvrB | |
| 6 | View Details | [1157..1269] | N/A | Confident ab initio structure predictions are available. | |
| 7 | View Details | [1270..1373] | 405.440336 | HRDC domain from RecQ helicase | |
| 8 | View Details | [1374..1447] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)