













| General Information: |
|
| Name(s) found: |
TCPB_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 527 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell wall
[IDA]
anchored to plasma membrane [IDA] |
| Biological Process: |
cellular protein metabolic process
[IEA]
protein folding [IEA] |
| Molecular Function: |
ATP binding
[IEA]
protein binding [IEA] unfolded protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPIDKIFKDD ASEEKGERAR MASFVGAMAI SDLVKSTLGP KGMDKILQST GRGHAVTVTN 60
61 DGATILKSLH IDNPAAKVLV DISKVQDDEV GDGTTSVVVL AGELLREAEK LVASKIHPMT 120
121 IIAGYRMASE CARNALLKRV IDNKDNAEKF RSDLLKIAMT TLCSKILSQD KEHFAEMAVD 180
181 AVFRLKGSTN LEAIQIIKKP GGSLKDSFLD EGFILDKKIG IGQPKRIENA NILVANTAMD 240
241 TDKVKIYGAR VRVDSMTKVA EIEGAEKEKM KDKVKKIIGH GINCFVNRQL IYNFPEELFA 300
301 DAGILAIEHA DFEGIERLGL VTGGEIASTF DNPESVKLGH CKLIEEIMIG EDKLIHFSGC 360
361 EMGQACSIVL RGASHHVLDE AERSLHDALC VLSQTVNDTR VLLGGGWPEM VMAKEVDELA 420
421 RKTAGKKSHA IEAFSRALVA IPTTIADNAG LDSAELVAQL RAEHHTEGCN AGIDVITGAV 480
481 GDMEERGIYE AFKVKQAVLL SATEASEMIL RVDEIITCAP RRREDRM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
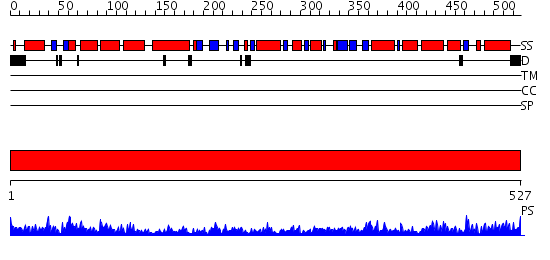
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..527] | 135.0 | Crystal structure of the chaperonin from Thermococcus strain KS-1 (nucleotide-free form of single mutant) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)