













| General Information: |
|
| Name(s) found: |
KAD5_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 588 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
chloroplast envelope [IDA] chloroplast thylakoid membrane [IDA] |
| Biological Process: |
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
[IEA]
|
| Molecular Function: |
ATP binding
[IEA]
adenylate kinase activity [IEA] nucleotide kinase activity [IEA][ISS] nucleobase, nucleoside, nucleotide kinase activity [IEA] phosphotransferase activity, phosphate group as acceptor [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASLSLSSAH FSSTSSSSRS SISTSSLSPS STSLPLLQSP IRRRYRSLRR RLSFSVIPRR 60
61 TSRSFSTSNS QIRCSINEPL KVMISGAPAS GKGTQCELIV HKFGLVHIST GDLLRAEVSS 120
121 GTDIGKRAKE FMNSGSLVPD EIVIAMVAGR LSREDAKEHG WLLDGFPRSF AQAQSLDKLN 180
181 VKPDIFILLD VPDEILIDRC VGRRLDPVTG KIYHIKNYPP ESDEIKARLV TRPDDTEEKV 240
241 KARLQIYKQN SEAIISAYSD VMVKIDANRP KEVVFEETQT LLSQIQLKRM IKTDKASPVQ 300
301 DKWRGIPTRL NNIPHSRDIR AYFYEDVLQA TIRSIKDGNT RLRVDINIPE LNPEMDVYRI 360
361 GTLMELVQAL ALSFADDGKR VKVCVQGSMG EGALAGMPLQ LAGTRKILEY MDWGDDETLG 420
421 TFVKLGAIGG KEVDEEDDMF ILVAPQNAVG NCIIDDLQAM TTAAGKRPVV LINPRLKDLP 480
481 ASSGIMQTMG REQRLEYALT FDNCYVFRLL YYLGTQYPIM GALRMSYPYR YELYKRVNEE 540
541 NGKEKYVLLA TYAERPTPEQ IDDAFSGKSR DQSKKASGIW GFLSSVFS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
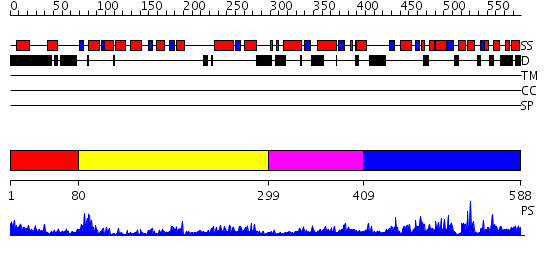
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..79] | 9.522879 | Signal sequence recognition protein Ffh; GTPase domain of the signal sequence recognition protein Ffh | |
| 2 | View Details | [80..298] | 46.0 | Adenylate kinase; Microbial and mitochondrial ADK, insert "zinc finger" domain | |
| 3 | View Details | [299..408] | 11.0 | Catalytic domain of E.coli Lon protease | |
| 4 | View Details | [409..588] | 31.585027 | No description for PF09353.1 was found. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|