













| General Information: |
|
| Name(s) found: |
MUTS_ECOBW
[Swiss-Prot]
|
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli BW2952 |
| Length: | 853 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSAIENFDAH TPMMQQYLRL KAQHPEILLF YRMGDFYELF YDDAKRASQL LDISLTKRGA 60
61 SAGEPIPMAG IPYHAVENYL AKLVNQGESV AICEQIGDPA TSKGPVERKV VRIVTPGTIS 120
121 DEALLQERQD NLLAAIWQDS KGFGYATLDI SSGRFRLSEP ADRETMAAEL QRTNPAELLY 180
181 AEDFAEMSLI EGRRGLRRRP LWEFEIDTAR QQLNLQFGTR DLVGFGVENA PRGLCAAGCL 240
241 LQYAKDTQRT TLPHIRSITM EREQDSIIMD AATRRNLEIT QNLAGGAENT LASVLDCTVT 300
301 PMGSRMLKRW LHMPVRDTRV LLERQQTIGA LQDFTAGLQP VLRQVGDLER ILARLALRTA 360
361 RPRDLARMRH AFQQLPELRA QLETVDSAPV QALREKMGEF AELRDLLERA IIDTPPVLVR 420
421 DGGVIASGYN EELDEWRALA DGATDYLERL EVRERERTGL DTLKVGFNAV HGYYIQISRG 480
481 QSHLAPINYM RRQTLKNAER YIIPELKEYE DKVLTSKGKA LALEKQLYEE LFDLLLPHLE 540
541 ALQQSASALA ELDVLVNLAE RAYTLNYTCP TFIDKPGIRI TEGRHPVVEQ VLNEPFIANP 600
601 LNLSPQRRML IITGPNMGGK STYMRQTALI ALMAYIGSYV PAQKVEIGPI DRIFTRVGAA 660
661 DDLASGRSTF MVEMTETANI LHNATEYSLV LMDEIGRGTS TYDGLSLAWA CAENLANKIK 720
721 ALTLFATHYF ELTQLPEKME GVANVHLDAL EHGDTIAFMH SVQDGAASKS YGLAVAALAG 780
781 VPKEVIKRAR QKLRELESIS PNAAATQVDG TQMSLLSVPE ETSPAVEALE NLDPDSLTPR 840
841 QALEWIYRLK SLV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
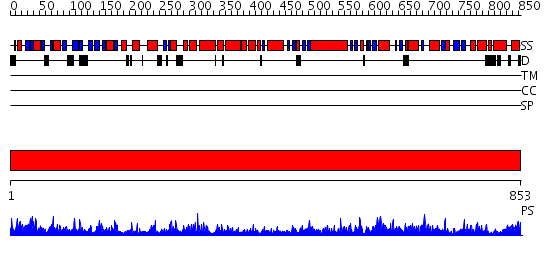
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..853] | 1000.0 | DNA repair protein MutS, domain III; DNA repair protein MutS, the C-terminal domain; DNA repair protein MutS, domain II; DNA repair protein MutS, domain I |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)