













| General Information: |
|
| Name(s) found: |
RCD1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 589 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][ISS]
cytoplasm [IDA] |
| Biological Process: |
embryonic development
[IMP]
response to salt stress [IMP] response to ozone [IMP] response to osmotic stress [IMP] ethylene mediated signaling pathway [IMP] response to superoxide [IMP] [NOT]response to hydrogen peroxide [IMP] jasmonic acid mediated signaling pathway [IMP] response to ethylene stimulus [IMP] response to water deprivation [IMP] response to oxidative stress [IGI] defense response to bacterium, incompatible interaction [IMP] nitric oxide biosynthetic process [IMP] programmed cell death [IMP] lateral root morphogenesis [IMP] |
| Molecular Function: |
protein binding
[IPI]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEAKIVKVLD SSRCEDGFGK KRKRAASYAA YVTGVSCAKL QNVPPPNGQC QIPDKRRRLE 60
61 GENKLSAYEN RSGKALVRYY TYFKKTGIAK RVMMYENGEW NDLPEHVICA IQNELEEKSA 120
121 AIEFKLCGHS FILDFLHMQR LDMETGAKTP LAWIDNAGKC FFPEIYESDE RTNYCHHKCV 180
181 EDPKQNAPHD IKLRLEIDVN GGETPRLNLE ECSDESGDNM MDDVPLAQRS SNEHYDEATE 240
241 DSCSRKLEAA VSKWDETDAI VVSGAKLTGS EVLDKDAVKK MFAVGTASLG HVPVLDVGRF 300
301 SSEIAEARLA LFQKQVEITK KHRGDANVRY AWLPAKREVL SAVMMQGLGV GGAFIRKSIY 360
361 GVGIHLTAAD CPYFSARYCD VDENGVRYMV LCRVIMGNME LLRGDKAQFF SGGEEYDNGV 420
421 DDIESPKNYI VWNINMNTHI FPEFVVRFKL SNLPNAEGNL IAKRDNSGVT LEGPKDLPPQ 480
481 LESNQGARGS GSANSVGSST TRPKSPWMPF PTLFAAISHK VAENDMLLIN ADYQQLRDKK 540
541 MTRAEFVRKL RVIVGDDLLR STITTLQNQP KSKEIPGSIR DHEEGAGGL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
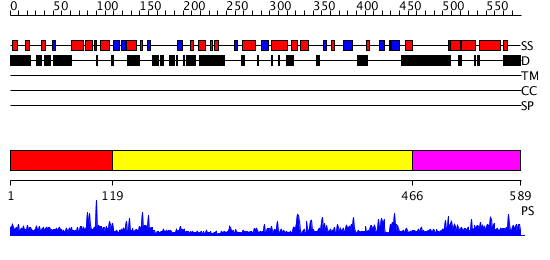
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..118] | 2.054982 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [119..465] | 56.154902 | THE CATALYTIC FRAGMENT OF POLY(ADP-RIBOSE) POLYMERASE COMPLEXED WITH CARBA-NAD | |
| 3 | View Details | [466..589] | 3.02898 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)