













| General Information: |
|
| Name(s) found: |
CCR4A_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 602 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
hydrolase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSVIRVHLP SEIPIVGCEL TPYVLLRRPD KTPSTDDVPE SAPLEGHFLK YRWFRVQSDK 60
61 KVAICSVHPS ETATLQCLGC LKSKVPVAKS YHCSTKCFSD AWQHHRVLHE RAASAATEGN 120
121 DEEELPRLNS SGSGSGVLST SVSLTNGSSS VYPSAITQKT GAGGETLVEV GRSKTYTPMA 180
181 DDICHVLKFE CVVVNAETKQ NVGLSCTILT SRVIPAPSPS PRRLISISGT DVTGHLDSNG 240
241 RPLSMGTFTV LSYNILSDTY ASSDIYSYCP TWALAWTYRR QNLLREIVKY RADIVCLQEV 300
301 QNDHFEEFFL PELDKHGYQG LFKRKTNEVF IGNTNTIDGC ATFFRRDRFS HVKKYEVEFN 360
361 KAAQSLTEAI IPVSQKKNAL NRLVKDNVAL IVVLEAKFGS QAADNPGKRQ LLCVANTHVN 420
421 VPHELKDVKL WQVHTLLKGL EKIAASADIP MLVCGDFNTV PASAPHTLLA VGKVDPLHPD 480
481 LMVDPLGILR PHSKLTHQLP LVSAYSQFAK MGGNVITEQQ RRRLDPASSE PLFTNCTRDF 540
541 IGTLDYIFYT ADTLTVESLL ELLDEESLRK DTALPSPEWS SDHIALLAEF RCMPRARRNN 600
601 IL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
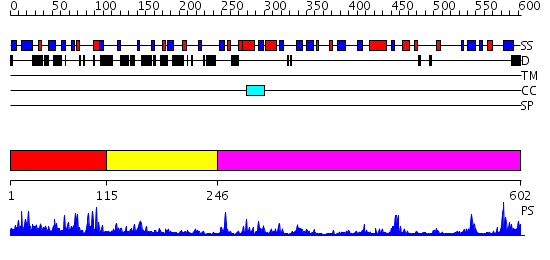
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..114] | 1.6 | No description for 2d8qA was found. | |
| 2 | View Details | [115..245] | 18.0 | Crystal structure of Human Methionine Aminopeptidase Type I with a third cobalt in the active site | |
| 3 | View Details | [246..602] | 27.0 | DNA repair endonuclease Hap1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)