













| General Information: |
|
| Name(s) found: |
DEAH4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 700 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
nucleotide binding
[IEA]
nucleic acid binding [IEA] ATP binding [IEA] ATP-dependent helicase activity [IEA] nucleoside-triphosphatase activity [IEA] helicase activity [IEA] ATP-dependent RNA helicase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MANLPILQFE EKIVETVEKN SVVVIIGETG SGKSTQLSQI LHRHGYTKSG VIAITQPRRV 60
61 AAVSVARRVA QELDVPLGED VGYAIRFEDR TTSKTRIKYL TDGVLLRESL SNPMLDDYSV 120
121 IILDEAHERS LNTDILLGLV KRLVRIRASN FKVLITSATL DGEKVSEFFS GCPVLNVPGK 180
181 LYPVEILYSK ERPVSYIESS LKVAIDIHVR EPEGDILIFM TGQDDIEKLV SRLEEKVRSL 240
241 AEGSCMDAII YPLHGSLPPE MQVRVFSPPP PNCRRFIVST NIAETSLTVD GVVYVIDSGY 300
301 VKQRQYNPSS GMFSLDVIQI SKVQANQRAG RAGRTRPGKC YRLYPLAVYR DDFLDATIPE 360
361 IQRTSLAGSV LYLKSLDLPD IDILKFDFLD APSSESLEDA LKQLYFIDAI DENGAITRIG 420
421 RTMSDLPLEP SLSRTLIEAN ETGCLSQALT VVAMLSAETT LLPARSKPSE KKRKHDEDSN 480
481 LPNGSGYGDH IQLLQIFESW DRTNYDPVWC KENGMQVRGM VFVKDVRRQL CQIMQKISKD 540
541 RLEVGADGRK SSSRDDYRKL RKALCVGNAN QIAERMLRHN GYRTLSFQSQ LVQVHPSSVL 600
601 SADNDGMMPN YVVYHELIST TRPFMRNVCA VDMAWVAPIK RKIEKLNVRK LSGGPAPSFK 660
661 VPEEKTELSK NNAETPAVSE NVESRIEAAR ERFLARKGQK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
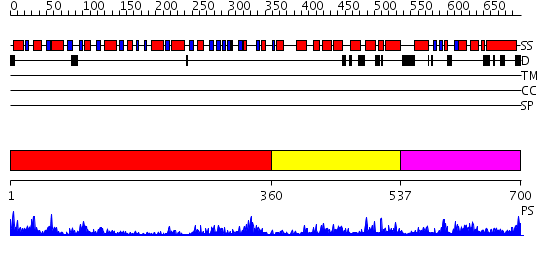
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..359] | 56.39794 | No description for 2hyiC was found. | |
| 2 | View Details | [360..536] | 1.3 | No description for 2v8oA was found. | |
| 3 | View Details | [537..700] | 23.136677 | No description for PF07717.7 was found. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)