













| General Information: |
|
| Name(s) found: |
gi|190404649
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae RM11-1a |
| Length: | 632 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKFTWKELI QLGSPSKAYE SSLACIAHID MNAFFAQVEQ MRCGLSKEDP VVCVQWNSII 60
61 AVSYAARKYG ISRMDTIQEA LKKCSNLIPI HTAVFKKGED FWQYHDGCGS WVQDPAKQIS 120
121 VEDHKVSLEP YRRESRKALK IFKSACDLVE RASIDEVFLD LGRICFNMLM FDNEYELTGD 180
181 LKLKDALSNI REAFIGGNYD INSHLPLIPE KIKSLKFEGD VFNPEGRDLI TDWDDVILAL 240
241 GSQVCKGIRD SIKDILGYTT SCGLSSTKNV CKLASNYKKP DAQTIVKNDC LLDFLDCGKF 300
301 EITSFWTLGG VLGKELIDVL DLPHENSIKH IRETWPDNAG QLKEFLDAKV KQSDYDRSTS 360
361 NIDPLKTADL AEKLFKLSRG RYGLPLSSRP VVKSMMSNKN LRGKSCNSIV DCISWLEVFC 420
421 AELTSRIQDL EQEYNKIVIP RTVSISLKTK SYEVYRKSGP VAYKGINFQS HELLKVGIKF 480
481 VTDLDIKGKN KSYYPLTKLS MTITNFDIID LQKTVVDMFG NQVHTFKSSA GKEDEEKTTS 540
541 SKADEKTPKL ECCKYQVTFT DQKALQEHAD YHLALKLSEG LNGAEESSKN LSFGEKRLLF 600
601 SRKRPNSQHT ATPQKKQVTS SKNILSFFTR KK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
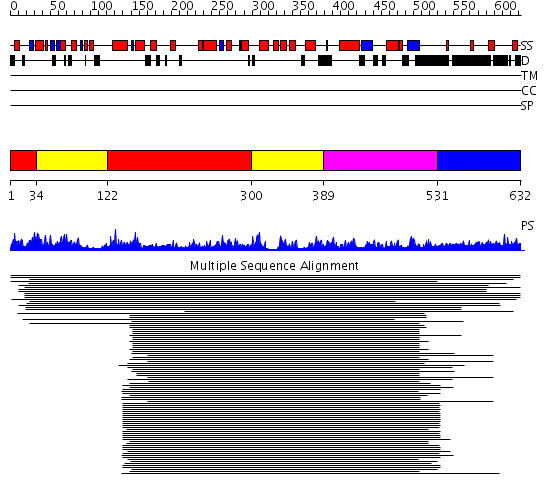
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..33] [122..299] |
10119.0 | DNA polymerase eta | |
| 2 | View Details | [34..121] [300..388] |
10119.0 | DNA polymerase eta | |
| 3 | View Details | [389..530] | 10119.0 | DNA polymerase eta | |
| 4 | View Details | [531..632] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)