













| General Information: |
|
| Name(s) found: |
gi|190407086
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae RM11-1a |
| Length: | 889 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKLDIKKTFS NRSDRVKGID FHPTEPWVLT TLYSGRVELW NYETQVEVRS IQVTETPVRA 60
61 GKFIARKNWI IVGSDDFRIR VFNYNTGEKV VDFEAHPDYI RSIAVHPTKP YVLSGSDDLT 120
121 VKLWNWENNW ALEQTFEGHE HFVMCVAFNP KDPSTFASGC LDRTVKVWSL GQSTPNFTLT 180
181 TGQERGVNYV DYYPLPDKPY MITASDDLTI KIWDYQTKSC VATLEGHMSN VSFAVFHPTL 240
241 PIIISGSEDG TLKIWNSSTY KVEKTLNVGL ERSWCIATHP TGRKNYIASG FDNGFTVLSL 300
301 GNDEPTLSLD PVGKLVWSGG KNAAASDIFT AVIRGNEEVE QDEPLSLQTK ELGSVDVFPQ 360
361 SLAHSPNGRF VTVVGDGEYV IYTALAWRNK AFGKCQDFVW GPDSNSYALI DETGQIKYYK 420
421 NFKEVTSWSV PMHSAIDRLF SGALLGVKSD GFVYFFDWDN GTLVRRIDVN AKDVIWSDNG 480
481 ELVMIVNTNS NGDEASGYTL LFNKDAYLEA ANNGNIDDSE GVDEAFDVLY ELSESITSGK 540
541 WVGDVFIFTT ATNRLNYFVG GKTYNLAHYT KEMYLLGYLA RDNKVYLADR EVHVYGYEIS 600
601 LEVLEFQTLT LRGEIEEAIE NVLPNVEGKD SLTKIARFLE GQEYYEEALN ISPDQDQKFE 660
661 LALKVGQLTL ARDLLTDESA EMKWRALGDA SLQRFNFKLA VEAFTNAHDL ESLFLLHSSF 720
721 NNKEGLVTLA KDAERAGKFN LAFNAYWIAG DIQGAKDLLI KSQRFSEAAF LGSTYGLGDD 780
781 AVNDIVTKWK ENLILNGKNT VSERVCGAEG LPGSSSSGDA QPLIDLDSTP APEQADENKE 840
841 AEVEDSEFKE SNSEAVEAEK KEEEAPQQQQ SEQQPEQGEA VPEPVEEES |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
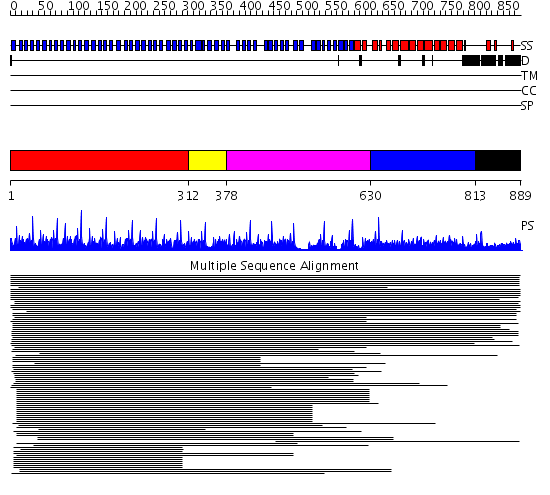
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..311] | 219.68867 | Tup1, C-terminal domain | |
| 2 | View Details | [312..377] | 3.106993 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [378..629] | 3.0 | TolB, C-terminal domain; TolB, N-terminal domain | |
| 4 | View Details | [630..812] | 10.2 | Clathrin heavy chain proximal leg segment | |
| 5 | View Details | [813..889] | 1.023991 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)