













| General Information: |
|
| Name(s) found: |
gi|207342631
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae AWRI1631 |
| Length: | 581 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSLASNTPL NGTPVSEAPA TSSEPVNMFE TMVANPIKVS RLQSNGVLTG PAANTKSIHY 60
61 SLANFNVFQS LPKETARGVD DLTRMEMALL SGIPEEIKWS LKKYLTYSNK APYMISLRTL 120
121 PDLLPLFKTF ILPLERIVEG LNKSSICDSK AMDSLQMGLN ALLILRNLAQ DTDSVQILVK 180
181 DREIKSFILF ILKKFQCVAT GDNKWQLYEG NATFFNELTH YTLDLMEAIS SYIAPAMKDD 240
241 HYFQTLVSIL NYTKDRYMVI SILRSLSRLL VRSKANEESA ADNLDHKTLS LIVSFLLLEC 300
301 DSELIIASLD FLYQYILPGS QRITELFKSK ECSLILEATL PNLLSYNIAT PDYHLLQKHK 360
361 IRLIKRLKPP APKEPPNLSE DLFQQLFKLN EPLRSTAWLR CCFEPVQEAE FTQISLWRSY 420
421 ESKFGQPVRE SGRKLLPAVE FIKNVSNAFN NAAAIVITDP VTGKKRFVIK GIQPRFKALG 480
481 IADGERESQV PISALKSKFL NDSKEITPAR QNSIPEVKFP QELSDVSKVA CTFLCLLSND 540
541 TDDGAGSAFC QRIRPLVLHK LADIPPLTLA LSEYMENTSG L |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
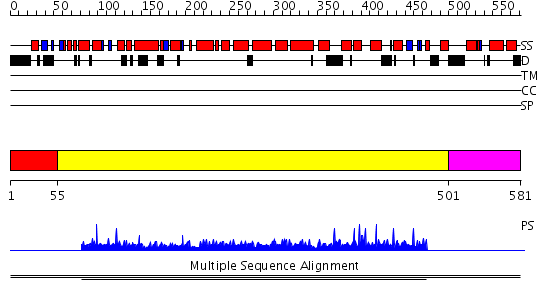
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..54] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [55..500] | 1.002847 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [501..581] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [482..581] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)