













| General Information: |
|
| Name(s) found: |
gi|205338197
[NCBI NR]
gi|168242649 [NCBI NR] |
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Salmonella enterica subsp. enterica serovar Heidelberg str. SL486 |
| Length: | 855 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNESFDKDFS NHTPMMQQYL KLKAQHPEIL LFYRMGDFYE LFYDDAKRAS QLLDISLTKR 60
61 GASAGEPIPM AGIPHHAVEN YLAKLVNQGE SVAICEQIGD PATSKGPVER KVVRIVTPGT 120
121 ISDEALLQER QDNLLAAIWQ DGKGYGYATL DISSGRFRLS EPADRETMAA ELQRTNPAEL 180
181 LYAEDFAEMA LIEGRRGLRR RPLWEFEIDT ARQQLNLQFG TRDLVGFGVE NASRGLCAAG 240
241 CLLQYVKDTQ RTSLPHIRSI TMERQQDSII MDAATRRNLE ITQNLAGGVE NTLAAVLDCT 300
301 VTPMGSRMLK RWLHMPVRNT DILRERQQTI GALQDTVSEL QPVLRQVGDL ERILARLALR 360
361 TARPRDLARM RHAFQQLPEL HAQLETVDSA PVQALRKKMG DFAELRDLLE RAIIDAPPVL 420
421 VRDGGVIAPG YHEELDEWRA LADGATDYLD RLEIRERERT GLDTLKVGYN AVHGYYIQIS 480
481 RGQSHLAPIN YVRRQTLKNA ERYIIPELKE YEDKVLTSKG KALALEKQLY DELFDLLLPH 540
541 LADLQQSANA LAELDVLVNL AERAWTLNYT CPTFTDKPGI RITEGRHPVV EQVLNEPFIA 600
601 NPLNLSPQRR MLIITGPNMG GKSTYMRQTA LIALLAYIGS YVPAQNVEIG PIDRIFTRVG 660
661 AADDLASGRS TFMVEMTETA NILHNATENS LVLMDEIGRG TSTYDGLSLA WACAENLANK 720
721 IKALTLFATH YFELTQLPEK MEGVANVHLD ALEHGDTIAF MHSVQDGAAS KSYGLAVAAL 780
781 AGVPKEVIKR ARQKLRELES ISPNAAATQV DGTQMSLLAA PEETSPAVEA LENLDPDSLT 840
841 PRQALEWIYR LKSLV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
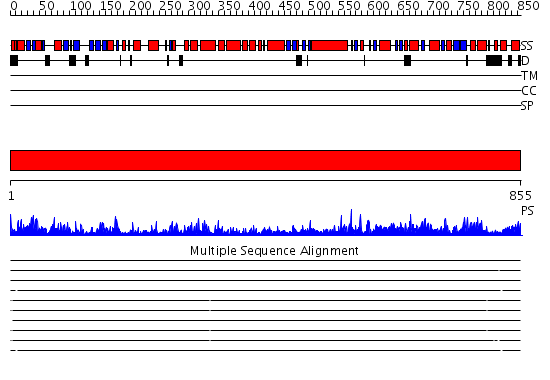
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..855] | 1000.0 | DNA repair protein MutS, domain III; DNA repair protein MutS, the C-terminal domain; DNA repair protein MutS, domain II; DNA repair protein MutS, domain I |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)