













| General Information: |
|
| Name(s) found: |
gi|213425494
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Salmonella enterica subsp. enterica serovar Typhi str. E02-1180 |
| Length: | 350 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQASQFSAQV LDWYDKYGRK TLPWQINKTP YKVWLSEVML QQTQVTTVIP YFERFMARFP 60
61 TVTDLANAPL DEVLHLWTGL GYYARARNLH KAAQQVATLH GGEFPQTFAE IAALPGVGRS 120
121 TAGAILSLAL GKHYPILDGN VKRVLARCYA VSGWPGKKEV ENTLWTLSEQ VTPARGVERF 180
181 NQAMMDLGAM VCTRSKPKCT LCPLQNGCIA AAHESWSRYP GKKPKQTLPE RTGYFLLLQH 240
241 NQEIFLAQRP PSGLWGGLYC FPQFASEDEL REWLAQRHVN ADNLTQLNAF RHTFSHFHLD 300
301 IVPMWLPVSS LDACMDEGGA LWYNLAQPPS VGLAAPVERL LQQLRTGAPV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
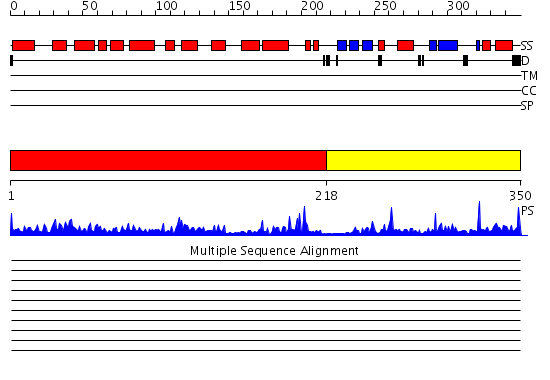
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..217] | 64.522879 | Catalytic domain of MutY | |
| 2 | View Details | [218..350] | 20.0 | NMR STUDY OF MUTT ENZYME, A NUCLEOSIDE TRIPHOSPHATE PYROPHOSPHOHYDROLASE |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)