













| General Information: |
|
| Name(s) found: |
MUKB_ECODH
[Swiss-Prot]
|
| Description(s) found:
Found 5 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli str. K-12 substr. DH10B |
| Length: | 1486 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIERGKFRSL TLINWNGFFA RTFDLDELVT TLSGGNGAGK STTMAAFVTA LIPDLTLLHF 60
61 RNTTEAGATS GSRDKGLHGK LKAGVCYSML DTINSRHQRV VVGVRLQQVA GRDRKVDIKP 120
121 FAIQGLPMSV QPTQLVTETL NERQARVLPL NELKDKLEAM EGVQFKQFNS ITDYHSLMFD 180
181 LGIIARRLRS ASDRSKFYRL IEASLYGGIS SAITRSLRDY LLPENSGVRK AFQDMEAALR 240
241 ENRMTLEAIR VTQSDRDLFK HLISEATNYV AADYMRHANE RRVHLDKALE FRRELHTSRQ 300
301 QLAAEQYKHV DMARELAEHN GAEGDLEADY QAASDHLNLV QTALRQQEKI ERYEADLDEL 360
361 QIRLEEQNEV VAEAIERQQE NEARAEAAEL EVDELKSQLA DYQQALDVQQ TRAIQYNQAI 420
421 AALNRAKELC HLPDLTADCA AEWLETFQAK ELEATEKMLS LEQKMSMAQT AHSQFEQAYQ 480
481 LVVAINGPLA RNEAWDVARE LLREGVDQRH LAEQVQPLRM RLSELEQRLR EQQEAERLLA 540
541 DFCKRQGKNF DIDELEALHQ ELEARIASLS DSVSNAREER MALRQEQEQL QSRIQSLMQR 600
601 APVWLAAQNS LNQLSEQCGE EFTSSQDVTE YLQQLLERER EAIVERDEVG ARKNAVDEEI 660
661 ERLSQPGGSE DQRLNALAER FGGVLLSEIY DDVSLEDAPY FSALYGPSRH AIVVPDLSQV 720
721 TEHLEGLTDC PEDLYLIEGD PQSFDDSVFS VDELEKAVVV KIADRQWRYS RFPEVPLFGR 780
781 AARESRIESL HAEREVLSER FATLSFDVQK TQRLHQAFSR FIGSHLAVAF ESDPEAEIRQ 840
841 LNSRRVELER ALSNHENDNQ QQRIQFEQAK EGVTALNRIL PRLNLLADDS LADRVDEIRE 900
901 RLDEAQEAAR FVQQFGNQLA KLEPIVSVLQ SDPEQFEQLK EDYAYSQQMQ RDARQQAFAL 960
961 TEVVQRRAHF SYSDSAEMLS GNSDLNEKLR ERLEQAEAER TRAREALRGH AAQLSQYNQV1020
1021 LASLKSSYDT KKELLNDLQR ELQDIGVRAD SGAEERARIR RDELHAQLSN NRSRRNQLEK1080
1081 ALTFCEAEMD NLTRKLRKLE RDYFEMREQV VTAKAGWCAV MRMVKDNGVE RRLHRRELAY1140
1141 LSADDLRSMS DKALGALRLA VADNEHLRDV LRMSEDPKRP ERKIQFFVAV YQHLRERIRQ1200
1201 DIIRTDDPVE AIEQMEIELS RLTEELTSRE QKLAISSRSV ANIIRKTIQR EQNRIRMLNQ1260
1261 GLQNVSFGQV NSVRLNVNVR ETHAMLLDVL SEQHEQHQDL FNSNRLTFSE ALAKLYQRLN1320
1321 PQIDMGQRTP QTIGEELLDY RNYLEMEVEV NRGSDGWLRA ESGALSTGEA IGTGMSILVM1380
1381 VVQSWEDESR RLRGKDISPC RLLFLDEAAR LDARSIATLF ELCERLQMQL IIAAPENISP1440
1441 EKGTTYKLVR KVFQNTEHVH VVGLRGFAPQ LPETLPGTDE APSQAS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
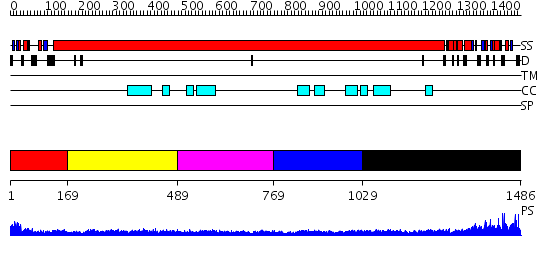
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..168] | 51.221849 | Cell division protein MukB | |
| 2 | View Details | [169..488] | 7.69897 | Tropomyosin | |
| 3 | View Details | [489..768] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [769..1028] | 4.522879 | No description for 2i1jA was found. | |
| 5 | View Details | [1029..1486] | 2.45 | Sc Smc1hd:Scc1-C complex, ATPgS |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)