













| General Information: |
|
| Name(s) found: |
gi|190404676
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae RM11-1a |
| Length: | 632 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MELSSNLKDL YIEWLQELVD GLTPKQEQLK IAYEKAKRNL QNAEGSFYYP TDLKKVKGIG 60
61 NTIIKRLDTK LRNYCKIHHI SPVEAPSLTQ TSSTRPPKRT TTALRSIVNS CENDKNEAPE 120
121 EKGTKKRKTR KYIPKKRSGG YAILLSLLEL NAIPRGVSKE QIIEVAGKYS DHCMTPNFST 180
181 KEFYGAWSSI AALKKHSLVL EEGRPKRYSL TEEGVELTKS LKTADGISFP KENEEPNEYS 240
241 VTRNESSEFT ANLTDLRGEY GKEEEPCDIN NTSFMLDITF QDLSTPQRLQ NNVFKNDRLN 300
301 SQTNISSHKL EEVSDDQTVP DSALKAKSTI KRRRYNGVSY ELWCSGDFEV FPIIDHREIK 360
361 SQSDREFFSR AFERKGMKSE IRQLALGDII WVAKNKNTGL QCVLNTIVER KRLDDLALSI 420
421 RDNRFMEQKN RLEKSGCEHK YYLIEETMSG NIGNMNEALK TALWVILVYY KFSMIRTCNS 480
481 DETVEKIHAL HTVISHHYSQ KDLIVIFPSD LKSKDDYKKV LLQFRREFER KGGIECCHNL 540
541 ECFQELMGKG DLKTVGELTI HVLMLVKGIS LEKAVAIQEI FPTLNKILMA YKTCSSEEEA 600
601 KLLMFNVLGD APGAKKITKS LSEKIYDAFG KL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
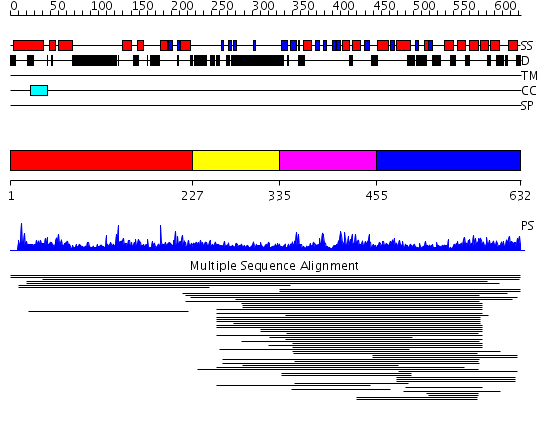
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..226] | 2.007945 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [227..334] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [335..454] | 50.086186 | ERCC4 domain No confident structure predictions are available. | |
| 4 | View Details | [455..632] | 3.038979 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)