













| General Information: |
|
| Name(s) found: |
TPS3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 730 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
trehalose biosynthetic process
[ISS]
|
| Molecular Function: |
alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGYDNVCGER QTLLVVANRL PASAKRTGEH SWSLEMSPGG KFNLLVEKDA VSKSLAEMKC 60
61 IPVFLNEVFD QYYNGYSNGI LWPILHHMGL PQEYDHDTIK TFETQYDAYK KANRMFLDVI 120
121 KENYKDGDIV WCQDYHLMFL PQYLKEYNNK IKVGWFLHSP FPSSEIYKTL PSRSELLRSV 180
181 LAADLISFHT YDFARHFVNT CTRILGVEGT HEGVVYQGRV TRVVVLPMGI YPNRFIKTCK 240
241 LPEVIQQMNE LKDRFSGKKV ILGVDRLDMI KGIPQKYLGF EKFLDENPNW RDKIVLVQIA 300
301 VPTRNEVPEY QKLKNQVHRL VGRINGRFGS VSSLPIHHMD CSVDSNYLCA LYAISDVMLV 360
361 TSLRDGLNLV SHEFVACQEA KRGVLILSEF AGAGQSLGAG ALLVNPWNVT EVSSAIKKAL 420
421 NMPYEERETR HRVNFKYVKT HSAEKWGFDF LSELNDAFDE SELQIRKIPH ELPQQDVIQR 480
481 YSLSNNRLII LNFGEYKIWL AAENGMFLKH TTEEWVTNMP QNMNLDWVDG LKNVFKYFTD 540
541 RTPRSFFEAS KTSLVWNYEY ADVEFGRAQA RDLLQYLWAG PISNASAEVV RGKYSVEVHA 600
601 IGVTKEPEIG HILGEIVHKK AMTTPIDYVF CSGYFLEKDE DIYTFFESEI LSPKLSHETR 660
661 SKSSSSNHSL EKKVSLNVLD LKQENYFSTA IGQARTKARY VVDSSHNVVN LLHKLAVANT 720
721 TTTSVKKPNV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
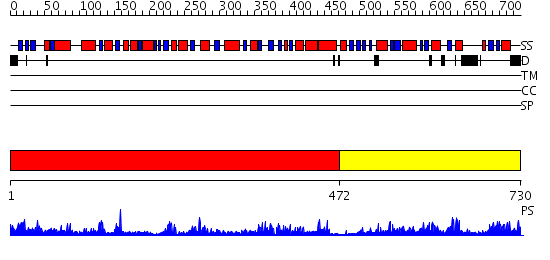
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..471] | 71.69897 | Trehalose-6-phosphate from E. coli bound with UDP-2-fluoro glucose. | |
| 2 | View Details | [472..730] | 6.69897 | X-Ray structure of the sucrose-phosphatase (SPP) from Synechocystis sp. PCC6803 at 1.40 A resolution |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)