













| General Information: |
|
| Name(s) found: |
gi|30685276
[NCBI NR]
gi|30685269 [NCBI NR] gi|110738501 [NCBI NR] gi|110737265 [NCBI NR] |
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 548 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
nucleic acid binding
[ISS]
RNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSPDHRMSPD HRDSHRKRSR PQSDYDDNGG SKRRYRGDDR DSLVIDRDDT VFRYLCPVKK 60
61 IGSVIGRGGD IVKQLRNDTR SKIRIGEAIP GCDERVITIY SPSDETNAFG DGEKVLSPAQ 120
121 DALFRIHDRV VADDARSEDS PEGEKQVTAK LLVPSDQIGC ILGRGGQIVQ NIRSETGAQI 180
181 RIVKDRNMPL CALNSDELIQ ISGEVLIVKK ALLQIASRLH ENPSRSQNLL SSSGGYPAGS 240
241 LMSHAGGPRL VGLAPLMGSY GRDAGDWSRP LYQPPRNDPP ATEFFIRLVS PVENIASVIG 300
301 KGGALINQLR QETRATIKVD SSRTEGNDCL ITISAREVFE DAYSPTIEAV MRLQPKCSDK 360
361 VERDSGLVSF TTRLLVPSSR IGCILGKGGA IITEMRRMTK ANIRILGKEN LPKVASDDDE 420
421 MVQISGELDV AKEALIQITS RLRANVFDRE GAVSALMPVL PYVPVAPDAG DRFDYDSRDS 480
481 RRLERGNPYP GGYGSSGVSA EGYSPYGAPV GGSSSTPYGV YGGYASGRSS SSGLSSHSST 540
541 YRRRNYDY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
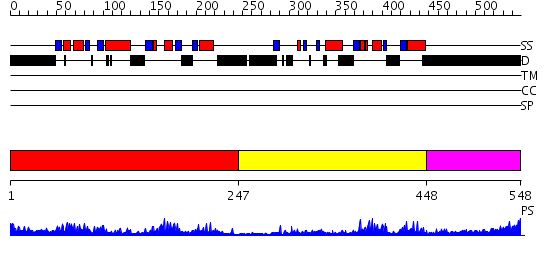
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..246] | 43.69897 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 | |
| 2 | View Details | [247..447] | 20.0 | Far upstream binding element, FBP, KH3 and KH4 domains | |
| 3 | View Details | [448..548] | 3.123996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)