













| General Information: |
|
| Name(s) found: |
AFG3 /
YER017C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 761 amino acids |
Gene Ontology: |
|
| Cellular Component: |
m-AAA complex
[IDA mitochondrion [IDA mitochondrial inner membrane [IDA |
| Biological Process: |
protein import into mitochondrial intermembrane space
[TAS protein complex assembly [IMP proteolysis [IMP signal peptide processing [IMP translation [IMP |
| Molecular Function: |
ATPase activity
[ISS metallopeptidase activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMMWQRYARG APRSLTSLSF GKASRISTVK PVLRSRMPVH QRLQTLSGLA TRNTIHRSTQ 60
61 IRSFHISWTR LNENRPNKEG EGKNNGNKDN NSNKEDGKDK RNEFGSLSEY FRSKEFANTM 120
121 FLTIGFTIIF TLLTPSSNNS GDDSNRVLTF QDFKTKYLEK GLVSKIYVVN KFLVEAELVN 180
181 TKQVVSFTIG SVDIFEEQMD QIQDLLNIPP RDRIPIKYIE RSSPFTFLFP FLPTIILLGG 240
241 LYFITRKINS SPPNANGGGG GGLGGMFNVG KSRAKLFNKE TDIKISFKNV AGCDEAKQEI 300
301 MEFVHFLKNP GKYTKLGAKI PRGAILSGPP GTGKTLLAKA TAGEANVPFL SVSGSEFVEM 360
361 FVGVGASRVR DLFTQARSMA PSIIFIDEID AIGKERGKGG ALGGANDERE ATLNQLLVEM 420
421 DGFTTSDQVV VLAGTNRPDV LDNALMRPGR FDRHIQIDSP DVNGRQQIYL VHLKRLNLDP 480
481 LLTDDMNNLS GKLATLTPGF TGADIANACN EAALIAARHN DPYITIHHFE QAIERVIAGL 540
541 EKKTRVLSKE EKRSVAYHEA GHAVCGWFLK YADPLLKVSI IPRGQGALGY AQYLPPDQYL 600
601 ISEEQFRHRM IMALGGRVSE ELHFPSVTSG AHDDFKKVTQ MANAMVTSLG MSPKIGYLSF 660
661 DQNDGNFKVN KPFSNKTART IDLEVKSIVD DAHRACTELL TKNLDKVDLV AKELLRKEAI 720
721 TREDMIRLLG PRPFKERNEA FEKYLDPKSN TEPPEAPAAT N |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
AFG3 YTA12 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
AFG3 YTA12 |
| View Details | Gavin AC, et al. (2006) |
|
AFG3 PHB2 YTA12 |
| View Details | Ho Y, et al. (2002) |
|
AFG3 ENP1 LPD1 |
| View Details | Qiu et al. (2008) |
|
AFG3 YTA12 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | 3829 - low filter | Green EM, et al (2005) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
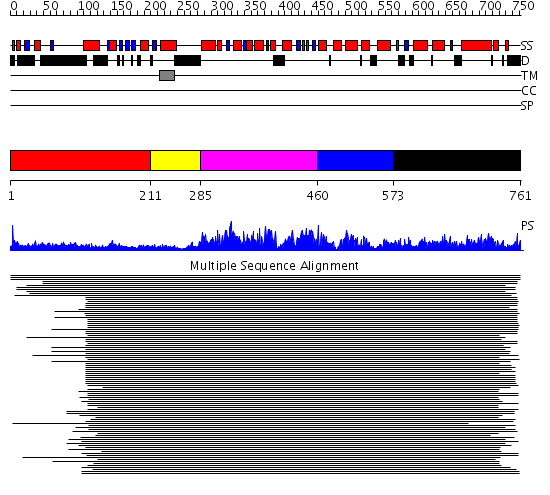
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..210] | 516.9897 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 2 | View Details | [211..284] | 516.9897 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 3 | View Details | [285..459] | 516.9897 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 4 | View Details | [460..572] | 516.9897 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 5 | View Details | [573..761] | 4.233982 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|