













| General Information: |
|
| Name(s) found: |
gi|55957427
[NCBI NR]
gi|55663742 [NCBI NR] gi|37620181 [NCBI NR] gi|24659293 [NCBI NR] gi|119574648 [NCBI NR] gi|119574646 [NCBI NR] |
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 1049 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLCWGNASFG QLGLGGIDEE IVLEPRKSDF FINKRVRDVG CGLRHTVFVL DDGTVYTCGC 60
61 NDLGQLGHEK SRKKPEQVVA LDAQNIVAVS CGEAHTLALN DKGQVYAWGL DSDGQLGLVG 120
121 SEECIRVPRN IKSLSDIQIV QVACGYYHSL ALSKASEVFC WGQNKYGQLG LGTDCKKQTS 180
181 PQLLKSLLGI PFMQVAAGGA HSFVLTLSGA IFGWGRNKFG QLGLNDENDR YVPNLLKSLR 240
241 SQKIVYICCG EDHTAALTKE GGVFTFGAGG YGQLGHNSTS HEINPRKVFE LMGSIVTEIA 300
301 CGRQHTSAFV PSSGRIYSFG LGGNGQLGTG STSNRKSPFT VKGNWYPYNG QCLPDIDSEE 360
361 YFCVKRIFSG GDQSFSHYSS PQNCGPPDDF RCPNPTKQIW TVNEALIQKW LSYPSGRFPV 420
421 EIANEIDGTF SSSGCLNGSF LAVSNDDHYR TGTRFSGVDM NAARLLFHKL IQPDHPQISQ 480
481 QVAASLEKNL IPKLTSSLPD VEALRFYLTL PECPLMSDSN NFTTIAIPFG TALVNLEKAP 540
541 LKVLENWWSV LEPPLFLKIV ELFKEVVVHL LKLYKIGIPP SERRIFNSFL HTALKVLEIL 600
601 HRVNEKMGQI IQYDKFYIHE VQELIDIRND YINWVQQQAY GMLADIPVTI CTYPFVFDAQ 660
661 AKTTLLQTDA VLQMQMAIDQ AHRQNVSSLF LPVIESVNPC LILVVRRENI VGDAMEVLRK 720
721 TKNIDYKKPL KVIFVGEDAV DAGGVRKEFF LLIMRELLDP KYGMFRYYED SRLIWFSDKT 780
781 FEDSDLFHLI GVICGLAIYN CTIVDLHFPL ALYKKLLKKK PSLDDLKELM PDVGRSMQQL 840
841 LDYPEDDIEE TFCLNFTITV ENFGATEVKE LVLNGADTAV NKQNRQEFVD AYVDYIFNKS 900
901 VASLFDAFHA GFHKVCGGKV LLLFQPNELQ AMVIGNTNYD WKELEKNTEY KGEYWAEHPT 960
961 IKIFWEVFHE LPLEKKKQFL LFLTGSDRIP ILGMKSLKLV IQSTGGGEEY LPVSHTCFNL1020
1021 LDLPKYTEKE TLRSKLIQAI DHNEGFSLI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
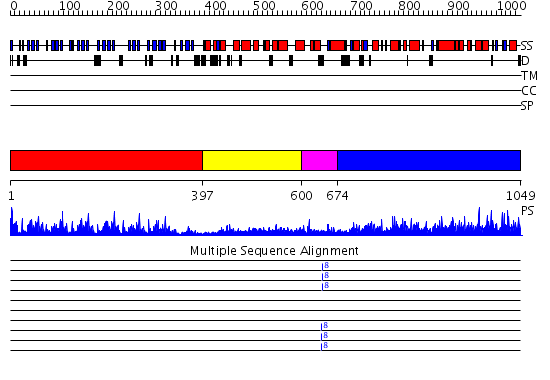
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..396] | 79.69897 | Regulator of chromosome condensation RCC1 | |
| 2 | View Details | [397..599] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [600..673] | 4.69897 | NMR structure of WW domains (WW3-4) from Suppressor of Deltex | |
| 4 | View Details | [674..1049] | 125.0 | Ubiquitin-protein ligase E3a, Hect catalytic domain (E6ap) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.42 |
Source: Reynolds et al. (2008)