













| General Information: |
|
| Name(s) found: |
YFIQ_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 6 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 886 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQRGLEALL RPKSIAVIGA SMKPNRAGYL MMRNLLAGGF NGPVLPVTPA WKAVLGVLAW 60
61 PDIASLPFTP DLAVLCTNAS RNLALLEELG EKGCKTCIIL SAPASQHEDL RACALRHNMR 120
121 LLGPNSLGLL APWQGLNASF SPVPIKRGKL AFISQSAAVS NTILDWAQQR KMGFSYFIAL 180
181 GDSLDIDVDE LLDYLARDSK TSAILLYLEQ LSDARRFVSA ARSASRNKPI LVIKSGRSPA 240
241 AQRLLNTTAG MDPAWDAAIQ RAGLLRVQDT HELFSAVETL SHMRPLRGDR LMIISNGAAP 300
301 AALALDALWS RNGKLATLSE ETCQKLRDAL PEHVAISNPL DLRDDASSEH YIKTLDILLH 360
361 SQDFDALMVI HSPSAAAPAT ESAQVLIEAV KHHPRSKYVS LLTNWCGEHS SQEARRLFSE 420
421 AGLPTYRTPE GTITAFMHMV EYRRNQKQLR ETPALPSNLT SNTAEAHLLL QQAIAEGATS 480
481 LDTHEVQPIL QAYGMNTLPT WIASDSTEAV HIAEQIGYPV ALKLRSPDIP HKSEVQGVML 540
541 YLRTANEVQQ AANAIFDRVK MAWPQARVHG LLVQSMANRA GAQELRVVVE HDPVFGPLIM 600
601 LGEGGVEWRP EDQAVVALPP LNMNLARYLV IQGIKSKKIR ARSALRPLDV AGLSQLLVQV 660
661 SNLIVDCPEI QRLDIHPLLA SGSEFTALDV TLDISPFEGD NESRLAVRPY PHQLEEWVEL 720
721 KNGERCLFRP ILPEDEPQLQ QFISRVTKED LYYRYFSEIN EFTHEDLANM TQIDYDREMA 780
781 FVAVRRIDQT EEILGVTRAI SDPDNIDAEF AVLVRSDLKG LGLGRRLMEK LITYTRDHGL 840
841 QRLNGITMPN NRGMVALARK LGFNVDIQLE EGIVGLTLNL AQREES |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
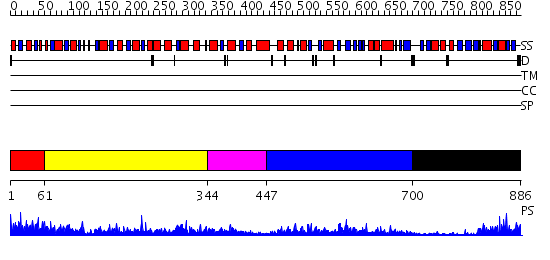
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..60] | 96.522879 | No description for 2csuA was found. | |
| 2 | View Details | [61..343] | 38.69897 | Succinyl-CoA synthetase, beta-chain, C-terminal domain; Succinyl-CoA synthetase, beta-chain, N-terminal domain | |
| 3 | View Details | [344..446] | 3.18 | No description for 2csuA was found. | |
| 4 | View Details | [447..699] | 32.69897 | Crystal structure of PH1788 from Pyrococcus horikoshii Ot3 | |
| 5 | View Details | [700..886] | 9.0 | The crystal structure of a probable N-acetyltransferase from Pseudomonas aeruginosa |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.70 |
Source: Reynolds et al. (2008)