













| General Information: |
|
| Name(s) found: |
SELB_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 614 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIIATAGHVD HGKTTLLQAI TGVNADRLPE EKKRGMTIDL GYAYWPQPDG RVPGFIDVPG 60
61 HEKFLSNMLA GVGGIDHALL VVACDDGVMA QTREHLAILQ LTGNPMLTVA LTKADRVDEA 120
121 RVDEVERQVK EVLREYGFAE AKLFITAATE GRGMDALREH LLQLPEREHA SQHSFRLAID 180
181 RAFTVKGAGL VVTGTALSGE VKVGDSLWLT GVNKPMRVRA LHAQNQPTET ANAGQRIALN 240
241 IAGDAEKEQI NRGDWLLADV PPEPFTRVIV ELQTHTPLTQ WQPLHIHHAA SHVTGRVSLL 300
301 EDNLAELVFD TPLWLADNDR LVLRDISARN TLAGARVVML NPPRRGKRKP EYLQWLASLA 360
361 RAQSDADALS VHLERGAVNL ADFAWARQLN GEGMRELLQQ PGYIQAGYSL LNAPVAARWQ 420
421 RKILDTLATY HEQHRDEPGP GRERLRRMAL PMEDEALVLL LIEKMRESGD IHSHHGWLHL 480
481 PDHKAGFSEE QQAIWQKAEP LFGDEPWWVR DLAKETGTDE QAMRLTLRQA AQQGIITAIV 540
541 KDRYYRNDRI VEFANMIRDL DQECGSTCAA DFRDRLGVGR KLAIQILEYF DRIGFTRRRG 600
601 NDHLLRDALL FPEK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
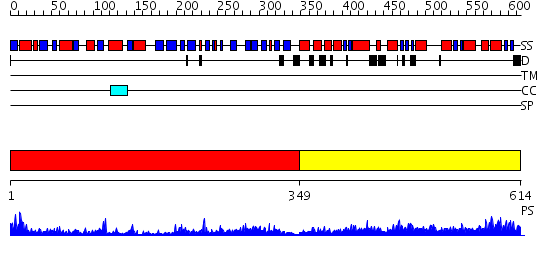
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..348] | 84.154902 | Elongation factor Tu (EF-Tu), domain 2; Elongation factor Tu (EF-Tu); Elongation factor Tu (EF-Tu), N-terminal (G) domain | |
| 2 | View Details | [349..614] | 48.154902 | C-terminal fragment of elongation factor SelB |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)