













| General Information: |
|
| Name(s) found: |
PURK_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 355 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKQVCVLGNG QLGRMLRQAG EPLGIAVWPV GLDAEPAAVP FQQSVITAEI ERWPETALTR 60
61 ELARHPAFVN RDVFPIIADR LTQKQLFDKL HLPTAPWQLL AERSEWPAVF DRLGELAIVK 120
121 RRTGGYDGRG QWRLRANETE QLPAECYGEC IVEQGINFSG EVSLVGARGF DGSTVFYPLT 180
181 HNLHQDGILR TSVAFPQANA QQQAQAEEML SAIMQELGYV GVMAMECFVT PQGLLINELA 240
241 PRVHNSGHWT QNGASISQFE LHLRAITDLP LPQPVVNNPS VMINLIGSDV NYDWLKLPLV 300
301 HLHWYDKEVR PGRKVGHLNL TDSDTSRLTA TLEALIPLLP PEYASGVIWA QSKFG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
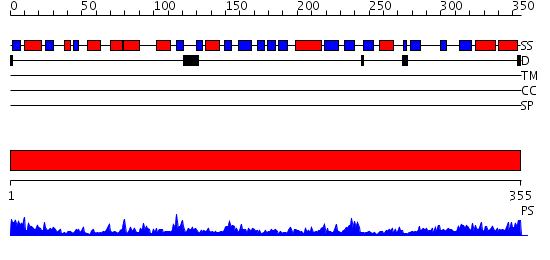
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..355] | 66.69897 | N5-carboxyaminoimidazole ribonucleotide synthetase PurK (AIRC), C-domain; N5-carboxyaminoimidazole ribonucleotide synthetase PurK (AIRC), N-domain; N5-carboxyaminoimidazole ribonucleotide synthetase PurK (AIRC), domain 2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)